Abstract
This study investigated the effects of seaweed extract (SWE) made from the brown algae Durvillaea potatorum and Ascophyllum nodosum on plants and soil. The application of SWE to soil growing tomato plants showed dual effects. SWE comprehensively improved tomato plant growth (flower clusters, flower number, fruit number, root length, root and shoot dry weight, SPAD) and increased plant productivity (yield and quality). Similarly, SWE application effected soil biology at the soil root zone by increasing total bacterial count and available soil nitrogen and impacting bacterial community diversity with an increase in certain bacterial families linked to soil health. A broader understanding of the effects of SWE on the plant-soil ecosystem may offer breakthrough approaches for sustainable food production.
Similar content being viewed by others
Introduction
The role of agricultural biostimulants in food production worldwide is expanding, especially those made from seaweed extracts (SWE). Biostimulants made from seaweeds have properties that improve plant growth, yield and quality and increase tolerance to abiotic and biotic stresses, as well as improving nutrient use (Arioli et al. 2015; Shukla et al. 2019). For example, field studies have demonstrated that SWE, applied at relatively low rates as foliar sprays or soil fertigation, increase yield among diverse crops such as wine grapes, sugarcane, strawberries and vegetables (Khan et al. 2009; Arioli et al. 2015; Shukla et al. 2019). Studies using Arabidopsis, soybean and tomato plants have demonstrated that the application of SWE can improve plant tolerance to abiotic stresses such as drought, freezing and salt (Nair et al. 2012; Martynenko et al. 2016; Santaniello et al. 2017; Goni et al. 2018; Jithesh et al. 2019). At a molecular level, laboratory studies have demonstrated that certain SWE have an intriguing capacity to activate plant defence response systems including the Systemic Acquired Resistance mechanism (Goni et al. 2016; Islam et al. 2020; Omidbakhshfard et al. 2020). In addition, soil-applied SWE provide a wide range of nutrients for soil microbiota utilisation.
The microbiota at the interface between plant roots and soil (rhizosphere) have been linked to improved plant growth and health (Vives-Peris et al. 2020). For example, microbiota at the rhizosphere can protect plants against pathogens (Mendes et al. 2011; Rolfe et al. 2019), help withstand stress to adverse environmental conditions (Rolli et al. 2015; Hartman and Tringe 2019) and alter plant development, for example, by influencing the timing of plant flowering (Lu et al. 2018; Olanrewaju et al. 2019). The rhizosphere environment is highly interactive because plants manipulate the composition of their root exudates (Olanrewaju et al. 2019). Consequently, a broader understanding of the effect of SWE on plant and soil biology is needed to advance agriculture.
We hypothesise that the soil application of a SWE made from two brown seaweeds (Ascophyllum nodosum and Durvillaea potatorum) would enhance plant growth and soil biology and would influence the plant-soil ecosystem. We chose an extract made from two seaweeds because transcriptomics research has shown an expanded set of plant gene responses compared with the extracts made from single-origin seaweeds (Islam et al. 2020). For new insights, we investigated tomato plant growth and soil biology parameters concurrently. We designed a greenhouse experiment using tomato plants growing in pots and receiving soil applications of SWE at regular intervals and scientific assessments focused on flowering and fruit harvest and quality. Soil biology was investigated using metagenomics sequencing at the commencement of flowering and phenotypic assessments followed for flowering, fruit number, root growth, leaf chlorophyll and fruit yield and quality. Flowering was chosen because the transition from vegetative to propagative growth is a critical plant development stage for field crops.
The aim of the study was to determine the effect of SWE concurrently on (i) plant growth and productivity and (ii) the rhizosphere microbiology.
Method
Seedling establishment
Tomato seeds (Lycopersicon esculentum Var. Grosse Lisse) were used in this study (from Seeds and Select Organic, Queensland, Australia). Commercial seedling trays were filled with commercially purchased seed raising mix (Debco Seed & Cutting Mix, Victoria, Australia). The tomato seeds were planted into the seedling trays at a rate of one seed per cell. The trays were placed at 21 °C for 4 weeks in a temperature-controlled greenhouse for germination and the soil mix was kept moist as required.
Experimental plant trial
A greenhouse potting trial was conducted using a commercially available seaweed extract manufactured by alkaline hydrolysis from Durvillaea potatorum and Ascophyllum nodosum and fortified with calcium at 1% (w/v) (Seasol Plus Calcium, Melbourne, Australia). Detailed information about the seaweed extract such as seaweed composition, mineral content and manufacture has been published previously (Arioli et al. 2015). A seaweed extract with calcium was selected because tomato fruits are susceptible to calcium disorders such as blossom end rot and yellow shoulder disorder. Four-week-old tomato seedlings were transplanted to pots (400 mm) containing Premium Potting Mix containing controlled-release fertiliser granules (N16%, P3%, K13%; Grow Better Garden Products, Victoria, Australia). Twenty pots containing the tomato seedlings were transferred into the greenhouse (8m × 8m × 6m, L × W × H) exposed to local ambient spring and summer conditions and not temperature controlled. The plants were climate hardened for a 1 week before the first treatment application. For application, the SWE was applied at 1:400 dilution and water was used for the untreated control. The SWE dilution rate was selected because of field and greenhouse studies demonstrating the efficacy at this dilution (Mattner et al. 2013, 2018; Arioli et al. 2020) and because of root growth testing (Arioli et al. 2015). The SWE treatment was applied to the soil in each pot using a piped irrigation system fitted with (i) a dosing unit (Dosatron, Australia) set to deliver the required dilution and (ii) a frequency controller set to deliver twice daily doses of 250 mL (to each pot). The trial was conducted as a randomised complete block design with ten replicates for each treatment, with 20 plants in total. Three plant developmental time points were used for the different assessments: flower initiation, flowering and harvest, which occurred at 29, 64 and 95 days after potting (DAP). The flower initiation day (29 DAP) was determined by monitoring the plants daily for the appearance of the first open flower.
Plant growth measurements
Plant growth was investigated at flowering and harvest using five replicates (plants) per treatment (ten plants in total) by assessing flower cluster and fruit number (Hussain et al. 2017), root length (Doerner 2008), shoot dry weight and root dry weight (Huang et al. 2017), and tomato leaf chlorophyll was assessed using a soil plant analysis development meter SPAD (SPAD-502, Konica Minolta, Japan) (Yao et al. 2020). Shoot and root samples were dried at 70 °C in a drying oven and dry weights were measured using analytical scales (Westlabs Model 663-543) after 72 h.
Soluble solids concentration
Tomato fruits harvested at 95 DAP were sent for fruit quality analysis (Freshhort, Victoria, Australia). Tomato fruit soluble solid concentration (SSC) in degree Brix (°Brix) were measured by cutting a ripe section from the cheek of each fruit within a replicate, placing all sections in a plastic bag, hand squeezing the sections and pouring 10 mL of extracted juice through a fine-mesh sieve into a 15-mL Eppendorf tube. Approximately 0.5 mL of juice was then poured from the tube onto the lens of a temperature-compensated digital refractometer (ATAGO PAL-1) with a measurement accuracy of ± 0.2 °Brix. The refractometer was calibrated with distilled water prior to SSC measurements of each replicate. The measurement was repeated, and the mean SSC in °Brix was calculated from the two measurements per replicate.
For each replicate, the remaining juice was frozen at – 20 °C. All juice samples were thawed at 20 °C, and 3 mL of juice from each sample was diluted in 5 mL of distilled water once the juice temperature in all samples was above 15 °C. The titratable acidity of each sample was then measured via endpoint titration to pH 8.2 with 0.1 M NaOH using an automatic titrator (Steroglass Titre X) and AS23 Micro autosampler. The mean titratable acidity for each replicate was calculated as grams of citric acid equivalent per litre of juice using the NaOH titre volume.
Soil testing
At flower initiation (29 DAP), the soil growing the potted tomato plants were assessed for microbiology using metagenomics sequencing, available soil nitrogen and the number of bacterial cells. The soil samples (approximately 400 g) were carefully collected from individual pots by selecting soil in contact with the central tomato root zone (rhizosphere) at a depth of 10 cm and avoiding the bulk soil zone (non-rhizosphere) that was not in contact with the roots. For metagenomics testing, ten individual soil samples were collected in total, five individual soil replicates for each treatment. The soil in contact with the roots (root zone soil) was placed into separate zip lock bags using a clean spatula. The soil samples, 50 g for each replicate, was sent to the Australian Genomic Research Foundation (AGRF) for metagenomics shotgun sequencing (NGS) and analysis. DNA was extracted from the soil using DNeasy PowerSoil Pro Kit (Qiagen, Germany). DNA libraries were prepared for each replicate using the Illumina TruSeq Nano DNA Library Prep Kit (Illumina, USA) with a 350 bp insert library and sequenced using the Illumina HiSeq 2500 platform (Illumina, USA) and 100 base pair (bp) paired read chemistry.
The root zone soil samples, containing 200g soil, five individual replicates per treatment, were sent for available soil nitrogen testing (SWEP Laboratories Pty Ltd, Melbourne, Australia). Available soil nitrogen refers to the nitrogen that is available for plants to absorb and measured as nitrate-nitrogen (NO3-N) only. The method involves a calcium chloride extraction and UV-visible colorimetric assay at 550 nm (Industry Method 7C2, Rayment and Lyons 2011).
The root zone soil samples, containing 50g soil, ten individual replicates per treatment, were sent for a standardised bacterial plate count test (SWEP Laboratories Pty Ltd, Melbourne, Australia). The test measures the number of bacterial colony forming units per gram of soil (cfu g−1).
Bioinformatics analysis
To build the taxonomical profiles from whole metagenomics sequencing dataset, the fastq files containing the raw reads were imported into CLC Genomics Workbench v20.0 (QIAGEN, Aarhus C Denmark, http://www.clcbio.com), using the following options: Illumina import, paired-reads, paired-end (forward-reverse) and minimum distance of 1 and a maximum distance of 1000 (default). Microbial Genomics Module (MGM) available as a plugin in the CLC Genomics Workbench was used to perform quality control (QC) and taxonomical profiling of the raw reads. The trimmed sequences were aligned to a CLC Microbial Reference Database (Version June 2019) from Genbank (250,000 basepairs). The permutational multivariate analysis of variance (PERMANOVA) test was conducted using CLC Genomics Workbench version v20.0 between two treatments based on Bray–Curtis distance among all taxonomic levels of each community. Beta diversity using principal coordinate analysis (PCoA) plots were generated among samples using CLC Microbial Genomics Module.
Statistical analysis
For the plant assessments the treatment effect on plants was analysed using a randomised block design experiment setup and Agronomic Research Management software (ARM, South Dakota, USA). GraphPad Prism statistical software (GraphPad, USA) was used to perform the statistical analysis. A t-test between the two treatments was used to show the statistical significance of alpha 0.05. Means followed by the same letter do not significantly differ. For the fruit quality (SSC) assessment, data for the treatment effect on fruit quality were analysed as a completely randomised design experiment using two-way ANOVA in GenStat 17 (VSN International Ltd., UK). Violations of the ANOVA assumption of normality in the data, such as non-normality (Skewness, Kurtosis) or heterogeneity of treatment variance, were assessed using residual error plots, skewness and kurtosis tests of normality and Bartlett’s test of homogeneity of variance. Where necessary, the appropriate data correction transformation was applied to data prior to ANOVA based on optimal values of lambda calculated from Box-Cox analysis in Genstat.
For the soil microbial analysis, at the family level, differential abundance analysis was done using the CLC Genomics Workbench v20.0. Post-QC and taxonomical profiling of the individual fastq files from each treatment, the CLC MGM tool was used to perform the differential abundance analysis between the two treatments. This CLC MGM tool performed a generalised linear model differential abundance test on the samples defined by metadata. The tool modelled each feature based on a separate Generalised Linear Model, where it assumed that the abundances followed a negative binomial distribution. The Wald test was used to determine significance between the treatments.
Results
Effects of SWE treatment on plant growth and productivity
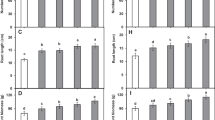
At the flowering (64 DAP) and harvest (95 DAP) stages, plant growth was assessed by determining (i) the number of flower clusters, (ii) fruit number, (iii) shoot dry weight and (iv) root dry weight. A statistical analysis found a significant increase in all the plant growth parameters in the SWE treatment relative to the untreated control (P < 0.05, Fig. 1; P = 0.0049 for day 64 DAP number of flower clusters, P = 0.0463 for day 95 DAP number of flower clusters, P = 0.0014 for day 95 DAP number of fruit, P = 0.0289 for day 64 DAP shoot dry weight, P = 0.0005 for 95 DAP shoot dry weight, P = 0.0149 for day 64 DAP root dry weight, P = 0.0207 for 95 DAP root dry weight) except for the average fruit number at the flowering stage. At flowering, the fruit number was found to be larger in the SWE-treated plants but not significantly.
The effect of SWE on the number of flower clusters, fruit number and shoot and root dry weight (64 DAP and 95 DAP). The SWE treatment significantly increased the four plant growth assessments relative to the water control. Black bars indicate SWE treatment, and white bars indicate untreated control. Means followed by the same letter do not significantly differ (P = 0.05, Fisher’s LSD; n = 5; means with standard error bars are indicated)
Additional plant assessments were undertaken at the flowering stage: (i) root length, (ii) flower number per plant and (iii) chlorophyll content. Statistical analysis found a significant increase in all the plant growth parameters due to the SWE treatment relative to the untreated control (P < 0.05, Fig. 2; P = 0.0475 for day 64 DAP root length, P = 0.0008 for day 64 DAP flower number per plant, P = 0.0220 for chlorophyll content).
The effect of SWE on flower number, SPAD and root length assessments at flowering (64 DAP). The SWE treatment significantly increased the three physiological assessments relative to the water control (untreated). Black bars indicate SWE treatment, and white bars indicate untreated control. Means followed by the same letter do not significantly differ (P = 0.05, Fisher’s LSD; n = 5; means with standard error bars are indicated)
Plant productivity was assessed by measuring fruit yield per plant. Statistical analysis found a significant increase in fruit weight per plant due to the SWE treatment relative to the untreated control (P = value < 0.05, Fig. 3; P = 0.0075 for day 95 DAP fruit weight per plant). Fruit quality was assessed by measuring SSC. Fruit harvested from SWE-treated plants had a mean SSC of 5.6 °Brix, and fruit from untreated plants had a mean SSC of 5.2 °Brix. Statistical analysis found that the increase (8%) in SSC due to the SWE treatment relative to the untreated control was significant at P < 0.1. The increases in the overall plant growth phenotypes are in alignment with productivity and quality increases.
The effect of SWE on fruit yield at harvest (95 DAP). The SWE treatment significantly increased fruit yield relative to the water control (Untreated). Black bar indicates SWE treatment, and white bar indicates untreated control. Means followed by the same letter do not significantly differ (P = 0.05, Fisher’s LSD; n = 5; means with standard error bars are indicated)
Soil microbiology and available soil nitrogen
At the flower initiation stage, soil collected at the tomato root zone were assessed for bacterial numbers and available soil nitrogen. Statistical analysis found a significant increase in the number of soil bacteria (P-value < 0.05, Fig. 4; P = 0.0097) and the available nitrogen in the soil (P-value < 0.05, Fig. 5; P = 0.0492) due to the SWE treatment relative to the untreated control. Also, we noted that the SWE treatment increased the total active fungi cfu g−1 (8.58 × 106 vs 5.97 × 106 ), Total active yeast cfu g−1 (1.02 × 105 vs 7.20 × 104) and total plant growth-promoting bacteria cfu g−1 (1.98 × 106 vs 1.36 × 106) by 2.2-, 1.4-, 1.4-fold, respectively, but these results were not statistically significant (at P < 0.05).
The effect of SWE on bacterial colony count (29 DAP). The SWE treatment significantly increased the colony count relative to the water control (untreated). Black bar indicates SWE treatment, and white bar indicates untreated control. Means followed by the same letter do not significantly differ (P = 0.05, Fisher’s LSD; n = 5; means with standard error bars are indicated)
Soil microbial composition
The root zone soils collected at the flower initiation stage were assessed for microbial composition. Taxonomic profiling was used to investigate the abundance profiles at the domain and microbial levels. For absolute abundance at the domain level, the average number of reads for both the treatments aligned to Bacteria (97.78%) followed by Eukaryota (2.03%) and Archaea (0.19%). At the microbial level, the stacked bar chart (Fig. 6) shows the relative abundance profiles after the alignment of the raw reads to the microbial reference database for the treatments (untreated control and SWE). The taxonomical profiling of the potting mix soil revealed 83 bacterial families between the treatments. Nakamurellaceae was the most abundant (38%) in the potting mix treated with SWE, and it was absent in the untreated control samples while Mycobacteriaceae, Bradyrhizobiaceae, Thermonosporaceae inter alia were common between the treatments. Differences in microbial diversity and abundance between the treatments were confirmed by additional statistical analysis.
Beta analysis—microbial communities cluster distinctly between treatments at flower initiation
The Bray-Curtis distance metric was used to determine multivariate sample distances, which were visualised through the ordination of a principal coordinate analysis (PCoA). The PCoA shows the control samples clustered relative to SWE treatment samples (S1). PCo1 was found to account for the majority of the variation (74%).
Differential abundance analysis between the treatments
Analysis using permutational multivariate analysis of variance (PERMANOVA) confirmed substantial differences in the structures of the bacterial communities between the SWE and control potting mix soils (P ≤ 0.05; Table 1). Among the bacterial taxonomic families detected, 38 complete bacterial taxonomic were significantly different between the SWE- and control treated potting mixes. The SWE treatment enriched 14 bacterial families that showed a log fold change greater than or equal to 2 (P ≤ 0.05; S2) and depleted 14 bacterial families that showed a log fold change lesser than or equal to − 2 (P ≤ 0.05; S3). Nakamurellaceae, Cytophagaceae Sterolibacteriaceae, Archangiaceae, Moraxellaceae, Brevibacteriaceae, Chlorobiaceae and Dermatophilaceae were the top 8 bacterial families significantly enriched in the potting mix soil treated with SWE (S2).
Discussion
The effect of SWE on plants
This study found that the effect of SWE on plants significantly increased numerous plant growth parameters: number of flower clusters and flowers, fruit number, dry weight of foliage and roots and root length. SWE had a positive effect on tomato plant yield and the total soluble solid concentration in tomato fruit. SWE application was found to significantly increase the chlorophyll levels in the tomato leaves at the flowering stage, as determined by SPAD testing, an assessment used to assess crop nitrogen status (Yuan et al. 2016; Gholizadeh et al. 2017). Collectively, the effects due to the SWE are agronomically significant as prerequisites for increasing tomato productivity and fruit quality.
In our study, it is unlikely that the plant responses are due to a fertiliser effect of the SWE since the experiment included fertiliser in the potting soil (in both the control and SWE treatment). The applied fertiliser was substantially higher than the nutrients in the diluted SWE that was used. Furthermore, field research using the same SWE used in this study found that repeated soil applications did not change the soil macronutrient content (Mattner et al. 2013).
Our results, using an extract made from dual seaweeds, are consistent with published research demonstrating that SWE can improve plant growth and productivity (Khan et al. 2009; Shukla et al. 2019). Tomato studies using SWE made from Ascophyllum nodosum or Sargassum horneri demonstrated these SWE can enhance different aspects of plant growth. For example, Ali et al. (2016) reported that an SWE made from A. nodosum increased the number of flower clusters and fruit yield in tomato greenhouse and field trials. Renaut et al. (2019) found an SWE made from A. nodosum increased fruit number, shoot dry weight and root dry weight. Yao et al. (2020) found that an SWE made from S. horneri increased tomato yield and enhanced SPAD, chlorophyll content and photosynthesis capacity. More broadly, the soil application of SWE made from dual seaweeds (A. nodosum and Durvillaea potatorum) has been reported to increase growth and yield parameters in broccoli, sugarcane and strawberry field trials (Mattner et al. 2013, 2018; Arioli et al. 2020). Santaniello et al. (2017) used another SWE to report the effects on photosynthetic performance in Arabidopsis. Islam et al. (2020) reported SWE increased root growth in Arabidopsis. Given the effects of SWE reported across many different farming and growth environments using different crops, this implies their mode of action is conserved with an inherent plasticity when used as a foliar spray or soil application.
The effect of SWE on Soil
We investigated the effect of SWE, as a soil application, on the soil microbiology at the onset of flowering and focused on the characterisation of the soil bacteria located at the rhizosphere rather than the bulk soil. Flowering is a critical transition development point where plants reallocate metabolism and require nutrition, especially potassium and calcium, for plant and fruit growth (Lee et al. 2017). In addition, Lu et al. (2018) reported the influence of rhizosphere microorganisms on the timing of plant flowering. Therefore, we utilised the flowering stage as an important juncture linking soil biology and plant productivity.
At flower initiation we found evidence for changes in the soil microbiology before the plant phenotype assessments were undertaken. Firstly, we found that the recurring applications of SWE increased the total number of bacteria in the soil. From an ecosystem perspective, the increase in soil bacteria is significant because the additional bacteria provide cellular matter to decompose and supply the soil carbon and nitrogen cycles. The SWE also increased the available nitrogen content in the soil, which highlights an uncharacterized influence on the bacterial communities that mineralize soil nitrogen. The differences in the soil bacterial communities are supported by a Bray-Curtis analysis confirming that the SWE treatment created diverse and distinct soil populations. Collectively, these soil changes are relevant to agriculture because nitrogen is an essential nutrient for plant growth, and the cellular matter is vital to the soil organic matter reservoir in depleted farming soils.
Our metagenome analysis revealed a plethora of microbial communities at the root-soil rhizosphere. Taxonomical profiling revealed three major microbial domains: Bacteria, Archaea and Eukaryota. The Bacteria domain dominated in relative abundance, compared with the Archaea and Eukaryota domains. Soils rich in nitrogen fertiliser have been found to harbour dominate bacterial populations (Jia and Conrad 2009; Monreal et al. 2018).
Differential abundance analysis found that the SWE treatment enhanced 14 bacterial families and reduced 14 bacterial families. Our results are in general agreement with reports that showed an increased number of bacterial families in soils treated with SWE (Khan et al. 2012, 2013; Shukla et al. 2019). Also, SWE has been shown to increase fungal families in soils amended with organic matter (Renaut et al. 2019).
Upon review of the 14 bacterial families that were significantly enriched by SWE treatment, we found certain members to have functions associated with enhanced soil properties and plant growth (Muhammad and Shahida 2006; Sgroy et al. 2009; Miransari 2013; Rolli et al. 2015; Shan et al. 2018; Rana et al. 2019). For example, some of the species that belong to the family Nakamurellaceae have been associated with symbiotic endophytes (Whitman 2015; Singh and Dubey 2018; Nouioui et al. 2019). The Cytophagaceae and Archangiaceae families have members known to digest polysaccharides for growth and for cycling carbon (Dawid 2000; McBride et al. 2014). The Sterolibacteriaceae family have members associated with enhancing plant tolerance to heavy metals (Chain et al. 2003). The families that were found to be reduced in abundance tended to be associated with marine habits or had no ascribable relevance except for the Beijerinkiaceae family due to their ability to grow in habits scarce in nitrogen. Although metagenomic analysis is a powerful technique, additional analysis is needed to undercover the functionalities of the communities and their fit within the agricultural ecosystem.
Effects of SWE at the ecosystem level
Our results demonstrate the application of SWE to soil-influenced plant growth and soil characteristics. However, plants and soils are part of a broader plant-growing ecosystem involving a multitude of interactions, natural nutrient cycles and an interrelated food web.
Plants growing in soils develop intimate associations with the soil microbial community, particularly at the rhizosphere (Hartman and Tringe 2019). These relationships can take multiple forms leading to beneficial, deadly or commensal interactions. Plants use their carbon-rich root exudes to influence their interactions in the ecosystem (Thakur and Geisen 2019). Plants can sequester microbial communities that facilitate plant nutrient uptake, the suppression of biotic stress, the activation plant defence responses and enhanced plant tolerance to abiotic pathogens (Olanrewaju et al. 2019; Rolfe et al. 2019; Vives-Peris et al. 2020).
Soil biogeochemistry includes an important role for microbes (Neal et al. 2020). Organic matter residing in soil is processed by microbes and acts to bind mineral and colloids particles. Microbes associated with these surfaces have distinct gene assemblages (Hartman and Tringe 2019; Neal et al. 2020). This type of research has led to the proposal that soil-microbe systems have self-organising capabilities with carbon flux being the critical factor in soil systems (Neal et al. 2020).
An important insight from our study is that the application of SWE to the soil will resonate at the ecosystem level when plants are growing. We hypothesise that plants with improved growth and root systems, due to SWE application, are supplying relatively more exudates into the soil, increasing microbial biomass, thereby benefiting the overall soil biogeochemical cycles. This proposal is consistent with research demonstrating that plant species with nutrient acquisitive properties regulate a plant-soil carbon-nitrogen feedback system operating at the soil root interface (Henneron et al. 2020; Wei et al. 2020). Despite the complexity, further research from the agroecosystem perspective is required to elucidate the collective benefits of using SWE for sustainable agriculture and food production.
References
Ali N, Farrell A, Ramsubhag A, Jayaraman J (2016) The effect of Ascophyllum nodosum extract on the growth, yield and fruit quality of tomato grown under tropical conditions. J Appl Phycol 28:1353–1362
Arioli T, Mattner S, Winberg P (2015) Applications of seaweed extracts in Australian agriculture: past, present and future. J Appl Phycol 27:2007–2015
Arioli T, Hepworth G, Farnsworth B (2020) Effect of seaweed extract application on sugarcane production. Proc Aust Soc Sugar Cane Technol 42:393–396
Chain P, Lamerdin J, Larimer F, Regala W, Lao V, Land M, Hauser L, Hooper A, Klotz M, Norton J, Sayavedra-Soto L, Arciero D, Hommes N, Whittaker M, Arp D (2003) Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J Bacteriol 185:2759–2773
Dawid W (2000) Biology and global distribution of myxobacteria in soils. FEMS Microbiol Rev 24:403–427
Doerner P (2008) Phenotypic analysis of Arabidopsis mutants: quantitative analysis of root growth. Cold Spring Harb Protoc 2008:prot4960
Gholizadeh A, Saberioon M, Boruvka L, Wayayok A, Soom M (2017) Leaf chlorophyll and nitrogen dynamics and their relationship to lowland rice yield for site-specific paddy management. Inf Process Agric 4:259–268
Goni O, Fort A, Quille P, Mckeown P, Spillane C, O’Connell S (2016) Comparative transcriptome analysis of two Ascophyllum nodosum extract biostimulants: same seaweed but different. J Agric Food Chem 64:2980–2989
Goni O, Quille P, O’Connell S (2018) Ascophyllum nodosum extract biostimulants and their role in enhancing tolerance to drought stress in tomato plants. Plant Physiol Biochem 126:63–73
Hartman K, Tringe S (2019) Interactions between plants and soil shaping the root microbiome under abiotic stress. Biochem J 476:2705–2724
Henneron L, Kardol P, Wardle D, Cros C, Fontaine S (2020) Rhizosphere control of soil nitrogen cycling: a key component of plant economic strategies. New Phytol 228:1269–1282
Huang P, de-Bashan L, Crocker T, Kloepper J, Bashan Y (2017) Evidence that fresh weight measurement is imprecise for reporting the effect of plant growth-promoting (rhizo)bacteria on growth promotion of crop plants. Biol Fertil Soils 53:199–208
Hussain I, Ahmed M, Khan S, Khalid A, Ali S, Hussain I, Sajid M (2017) Screening of different exotic lines of tomato (Lycopersicon esculentum L.) under the agro-climatic condition of Haripur. Pure Appl Biol 6:1251–1259
Islam M, Gan H, Ziemann M, Hussain H, Arioli T, Cahill D (2020) Phaeophyceaean (brown algal) extracts activate plant defense systems in Arabidopsis thaliana challenged with Phytophthora cinnamomi. Front Plant Sci 11:852
Jia Z, Conrad R (2009) Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol 11:1658–1671
Jithesh M, Shukla S, Kant P, Joshi J, Critchley T, Prithiviraj B (2019) Physiological and transcriptomics analyses reveal that Ascophyllum nodosum extracts induce salinity tolerance in Arabidopsis by regulating the expression of stress responsive genes. J Plant Growth Regul 38:463–478
Khan W, Rayirath U, Subramanian S, Jithesh M, Rayorath P, Hodges D, Critchley A, Norrie J, Prithiviraj B (2009) Seaweed extracts as biostimulants of plant growth and development. J Plant Growth Regul 28:386–399
Khan W, Zhai R, Souleimanov A, Critchley A, Smith D, Prithiviraj B (2012) Commercial extract of Ascophyllum nodosum improves root colonization of Alfalfa by its bacterial symbiont Sinorhizobium meliloti. Commun Soil Sci Plant Anal 43:2425–2436
Khan W, Palanisamy R, Critchley A, Smith D, Papadopoulos Y, Prithiviraj B (2013) Ascophyllum nodosum extract and its organic fractions stimulate Rhizobium root nodulation and growth of Medicago sativa (Alfalfa). Commun Soil Sci Plant Anal 44:900–908
Lee J, Rahman A, Azam H, Kim H, Kwon J (2017) Characterizing nutrient uptake kinetics for efficient crop production during Solanum lycopersicum var. cerasiforme Alef. growth in a closed indoor hydroponic system. PLoS One 12:e0177041
Lu T, Ke M, Lavoie M, Xiaoji F, Zhenyan Z, Zhengwei F, Liwei S, Gillings M, Penuelas P, Haifeng Q, Yong-Guan Z (2018) Rhizosphere microorganisms can influence the timing of plant flowering. Microbiome 6:231
Martynenko A, Shotton K, Astatkie T, Petrash G, Fowler C, Neily W, Critchley T (2016) Thermal imaging of soybean response to drought stress: the effect of Ascophyllum nodosum seaweed extract. SpringerPlus 5:1393
Mattner S, Wite D, Riches D, Porter I, Arioli T (2013) The effect of kelp extract on seedling establishment of broccoli on contrasting soil types in southern Victoria, Australia. Biol Agric Hortic 29:258–270
Mattner S, Milinkovic M, Arioli T (2018) Increased growth response of strawberry roots to a commercial extract from Durvillaea potatorum and Ascophyllum nodosum. J Appl Phycol 30:2943–2951
McBride M, Liu W, Lu X, Zhu Y, Zhang W (2014) The Family Cytophagaceae. In: Rosenberg E, DeLong E.F, Lory S, Stackebrandt E, Thompson F (eds) The Prokaryotes. Springer, Berlin pp 577-593.
Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider H, Piceno M, DeSantis Z, Andersen L, Bakker A, Raaijmakers M (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332:1097–1100
Miransari M (2013) Soil microbes and the availability of soil nutrients. Acta Physiol Plant 35:3075–3084
Monreal C, Zhang J, Koziel S, Vidmar J, González M, Matus F, Baxi S, Wu S, DeRosa M, Etcheverria P (2018) Bacterial community structure associated with the addition of nitrogen and the dynamics of soluble carbon in the rhizosphere of canola (Brassica napus) grown in a Podzol. Rhizosphere 5:16–25
Muhammad F, Shahida H (2006) Plant growth promotion by Brevibacterium under chromium stress. Res J Bot 1:24–29
Nair P, Kandasamy S, Zhang J, Ji X, Kirby C, Benkel B, Hodges M, Critchley A, Hitz D, Prithiviraj B (2012) Transcriptional and metabolomic analysis of Ascophyllum nodosum mediated freezing tolerance in Arabidopsis thaliana. BMC Genomics 13:643
Neal A, Bacq-Labreuil A, Zhang X, Clark I, Coleman K, Mooney S, Ritz K, Crawford J (2020) Soil as an extended composite phenotype of the microbial metagenome. Sci Rep 10:10649
Nouioui I, Cortés-Albayay C, Carro L, Castro J, Gtari M, Ghodhbane-Gtari F, Klenk H, Tisa L, Sangal V, Goodfellow M (2019) Genomic insights into plant-growth-promoting potentialities of the genus Frankia. Front Microbiol 10:457
Olanrewaju O, Ayangbenro A, Glick B, Babalola O (2019) Plant health: feedback effect of root exudates-rhizobiome interactions. Appl Microbiol Biotechnol 103:1155–1166
Omidbakhshfard A, Sujeeth N, Gupta S, Omranian N, Guinan J, Brotman Y, Nikoloski Z, Fernie A, Mueller-Roeber B, Gechev T (2020) A Biostimulant obtained from the seaweed Ascophyllum nodosum protects Arabidopsis thaliana from severe oxidative stress. Int J Mol Sci 21:474
Rana N, Khadka S, Marasini B, Joshi B, Poudel P, Khanal S, Parajuli N (2019) Isolation and characterization of soil myxobacteria from Nepal. J Instr Sci Technol 24:2467–9240
Rayment G, Lyons D (2011) Soil Chemical Methods - Australasia. CSIRO Publishing, Victoria
Renaut S, Masse J, Norrie J, Blal B, Hijri M (2019) A commercial seaweed extract structured microbial communities associated with tomato and pepper roots and significantly increased crop yield. Microb Biotechnol 12:1346–1358
Rolfe S, Griffiths J, Ton J (2019) Crying out for help with root exudates: adaptive mechanisms by which stressed plants assemble health-promoting soil microbiomes. Curr Opin Microbiol 49:73–82
Rolli E, Marasco R, Vigani G, Ettoumi B, Mapelli F, Deangelis M, Gandolfi C, Casati E, Previtali F, Gerbino R, Pierotti CF, Borin S, Sorlini C, Zocchi G, Daffonchio D (2015) Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait. Environ Microbiol 17:316–331
Santaniello A, Scartazza A, Gresta F, Loreti E, Biasone A, Tommaso D, Piaggesi A, Perata P (2017) Ascophyllum nodosum seaweed extract alleviates drought stress in Arabidopsis by affecting photosynthetic performance and related gene expression. Front Plant Sci 8:1362
Sgroy V, Cassán F, Masciarelli O, Del Papa M, Lagares A, Luna V (2009) Isolation and characterization of endophytic plant growth-promoting (PGPB) or stress homeostasis-regulating (PSHB) bacteria associated to the halophyte Prosopis strombulifera. Appl Microbiol Biotechnol 85:371–381
Shan W, Zhou Y, Liu H, Yu X (2018) Endophytic actinomycetes from tea plants (Camellia sinensis): Isolation, abundance, antimicrobial, and plant-growth-promoting activities. BioMed Res Int 1470305.
Shukla P, Mantin E, Adil M, Bajpai S, Critchley A, Prithiviraj B (2019) Ascophyllum nodosum-based biostimulants: sustainable applications in agriculture for the stimulation of plant growth, stress tolerance, and disease management. Front Plant Sci 10:655
Singh R, Dubey A (2018) Diversity and applications of endophytic actinobacteria of plants in special and other ecological niches. Front Microbiol 9:1767
Thakur M, Geisen S (2019) Trophic regulations of the soil microbiome. Trends Microbiol 27:771–780
Vives-Peris V, de Ollas C, Gómez-Cadenas A, Pérez-Clemente R (2020) Root exudates: from plant to rhizosphere and beyond. Plant Cell Rep 39:3–17
Wei X, Wang X, Cao P, Gao Z, Chen A, Han J (2020) Microbial community changes in the rhizosphere soil of healthy and rusty Panax ginseng and discovery of pivotal fungal genera associated with rusty roots. Biomed Res Int 8018525
Whitman WB (2015) Bergey's Manual of Systematics of Archaea and Bacteria. Wiley, New Jersey
Yao Y, Wang X, Chen B, Zhang M, Ma J (2020) Seaweed extract improved yields, leaf photosynthesis, ripening time, and net returns of tomato (Solanum lycopersicum Mill.). ACS Omega 5:4242–4249
Yuan Z, Cao Q, Zhang K, Ata-Ul-Karim Karim S, Tian Y, Zhu Y, Cao W, Liu X (2016) Optimal leaf positions for SPAD meter measurement in rice. Front Plant Sci 7:719
Acknowledgements
We recognise and thank Jeffery Lang for his excellent technical assistance growing the experimental plants and conducting the greenhouse assessments.
Funding
Funding for the experimental research was provided by Seasol International Pty Ltd (Australia). No funding was received to assist with the preparation or submission of this manuscript.
Author information
Authors and Affiliations
Contributions
All co-authors contributed to the final version of the work and approved the manuscript for publication. HH: leading the experimental research, interpretation of data and writing the manuscript; TA: interpretation of data and writing the manuscript; NK: bioinformatics and review of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Seasol International (SI) is the manufacturer of seaweed extract (Seasol) in Australia. TA and HH are scientific researchers at Seasol International. TA is an adjunct Associate Professor at Deakin University. The authors declare that the research was conducted in the absence of any commercial or financial relationship that could be construed as a potential conflict of interest.
Consent for publication
All the authors provide consent to publish.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
S1
. Principal coordinates analysis (PCoA) of microbial communities of potting mix at flower initiation. PCoA based on Bray–Curtis dissimilarity between untreated and SWE treatment. (DOCX 29 kb)
S2
. Differential Abundance Analysis showing the increased bacterial families due to the SWE treatment. (DOCX 15 kb)
S3
. Differential Abundance Analysis showing the decreased bacterial families due to SWE treatment (DOCX 15 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hussain, H.I., Kasinadhuni, N. & Arioli, T. The effect of seaweed extract on tomato plant growth, productivity and soil. J Appl Phycol 33, 1305–1314 (2021). https://doi.org/10.1007/s10811-021-02387-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-021-02387-2