Abstract
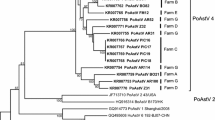
Noroviruses (NoVs) and sapoviruses (SaVs), which belong to the family Caliciviridae, are important human and animal enteric pathogens with zoonotic potential. In Ethiopia, no study has been done on the epidemiology of animal NoVs and SaVs. The aim of this study was to detect and characterize NoVs and SaVs from swine of various ages. Swine fecal samples (n = 117) were collected from commercial farms in Ethiopia. The samples were screened for caliciviruses by reverse transcription polymerase chain reaction (RT-PCR) using universal and genogroup-specific primer pairs. Phylogenetic analysis was conducted using a portion of the RNA-dependent RNA polymerase (RdRp) region and the VP1 region of genome sequences of caliciviruses. Among 117 samples, potential caliciviruses were detected by RT-PCR in 17 samples (14.5 %). Of the RT-PCR-positive fecal samples, four were sequenced, of which two were identified as human NoV GII.1 and the other two as porcine SaV GIII. The porcine SaV strains that were detected were genetically related to the porcine enteric calicivirus Cowden strain genogroup III (GIII), which is the prototype porcine SaV strain. No porcine NoVs were detected. Our results showed the presence of NoVs in swine that are most similar to human strains. These findings have important implications for NoV epidemiology and food safety. Therefore, continued surveillance of NoVs in swine is needed to define their zoonotic potential, epidemiology and public and animal health impact. This is the first study to investigate enteric caliciviruses (noroviruses and sapoviruses) in swine in Ethiopia.


Similar content being viewed by others
References
Saif LJ, Bohl EH, Theil KW, Cross RF, House JA (1980) Rotavirus-like, calicivirus-like, and 23-nm virus-like particles associated with diarrhea in young pigs. J Clin Microbiol 12(1):105–111
Wang QH, Han MG, Funk JA, Bowman G, Janies DA, Saif LJ (2005) Genetic diversity and recombination of porcine sapoviruses. J Clin Microbiol 43:5963–5972
Yin Y, Tohya Y, Ogawa Y, Numazawa D, Kato K, Akashi H (2006) Genetic analysis of calicivirus genomes detected in intestinal contents of piglets in Japan. Arch Virol 151(9):1749–1759
Nakamura K, Saga Y, Iwai M, Obara M, Horimoto E, Hasegawa S, Kurata T, Okumura H, Nagoshi M, Takizawa T (2010) Frequent detection of noroviruses and sapoviruses in swine and high genetic diversity of porcine sapovirus in Japan during fiscal year 2008. J Clin Microbiol 48:1215–1222
Kim HJ, Cho HS, Cho KO, Park NY (2006) Detection and molecular characterization of porcine enteric calicivirus in Korea, genetically related to sapoviruses. J Vet Med B Infect Dis Vet Public Health 53(4):155–159
Reuter G, Zimsek-Mijovski J, Poljsak-Prijatelj M, Di Bartolo I, Ruggeri FM, Kantala T, Maunula L, Kiss I, Kecskemeti S, Halaihel N, Buesa J, Johnsen C, Hjulsager CK, Larsen LE, Koopmans M, Bottiger B (2010) Incidence, diversity, and molecular epidemiology of sapoviruses in swine across Europe. J Clin Microbiol 48(2):363–368
L’Homme Y, Brassard J, Ouardani M, Gagne MJ (2010) Characterization of novel porcine sapoviruses. Arch Virol 155(6):839–846
Song YJ, Yu JN, Nam HM, Bak HR, Lee JB, Park SY, Song CS, Seo KH, Choi IS (2011) Identification of genetic diversity of porcine norovirus and sapovirus in Korea. Virus Genes 42(3):394–401
Dufkova L, Scigalkova I, Moutelikova R, Malenovska H, Prodelalova J (2013) Genetic diversity of porcine sapoviruses, kobuviruses, and astroviruses in asymptomatic pigs: an emerging new sapovirus GIII genotype. Arch Virol 158(3):549–558
Clarke IN, Lambden PR (2000) Organization and expression of calicivirus genes. J Infect Dis 181(Suppl 2):S309–S316
Atmar RL, Estes MK (2001) Diagnosis of non-cultivatable gastroenteritis viruses, the human caliciviruses. Clin Microbiol Rev 14(1):15–37
Green KY (2013) Caliciviridae: the noroviruses. In: Knipe DM, Howley PM (eds) Fields virology, 6thed, vol1. Lippincott Williams & Wilkins, Philadelphia, pp 583–609
Zheng DP, Ando T, Fankhauser RL, Beard RS, Glass RI, Monroe SS (2006) Norovirus classification and proposed strain nomenclature. Virology 346:312–323
Vega E, Barclay L, Gregoricus N, Shirley SH, Lee D, Vinjé J (2014) Genotypic and Epidemiologic Trends of Norovirus Outbreaks in the United States, 2009 to 2013. J Clin Microbiol 52(1):147–155
Wang QH, Han MG, Cheetham S, Souza M, Funk JA, Saif LJ (2005) Porcine noroviruses related to human noroviruses. Emerg Infect Dis 11(12):1874–1881
Costantini V, Fabienne L, Lynn J, Francoise S, Guyader Le, Saif LJ (2006) Human and animal enteric caliciviruses in Oysters from different coastal regions of the United States. Appl Env Microbiol 72(3):1800–1809
Phan TG, Okame M, Nguyen TA, Maneekarn N, Nishio O, Okitsu S (2007) Human astrovirus, norovirus (GI, GII), and sapovirus infections in Pakistani children with diarrhea. J Med Virol 79:633–638
Mattison K, Shukla A, Cook A, Pollari F, Friendship R, Kelton D, Bidawid S, Farber JM (2007) Human noroviruses in swine and cattle. Emerg Infect Dis 13:1184–1188
Farkas T, Zhong WM, Jing Y, Huang PW, Espinosa SM, Martinez N, Morrow AL, Ruiz-Palacios GM, Pickering LK, Jiang X (2004) Genetic diversity among sapoviruses. Arch Virol 149:1309–1323
Oka T, Mori K, Iritani N, Harada S, Ueki Y, Iizuka S, Mise K, Murakami K, Wakita T, Katayama K (2012) Human sapovirus classification based on complete capsid nucleotide sequences. Arch Virol 157(2):349–352
Sisay Z, Wang QH, Oka T, Saif LJ (2013) Prevalence and molecular characterization of porcine enteric caliciviruses and first detection of porcine kobuviruses in US swine. Arch Virol 158(7):1583–1588
Scheuer AK, Oka T, Hoet AE, Gebreyes WA, Molla BZ, Saif LJ, Wang QH (2013) Prevalence of porcine Noroviruses, molecular characterization of emerging porcine Sapoviruses from finisher swine in the United States, and unified classification scheme for Sapoviruses. J Clin Microbiol 51(7):2344–2353
Guo M, Chang KO, Hardy ME, Zhang Q, Parwani AV, Saif LJ (1999) Molecular characterization of a porcine enteric calicivirus genetically related to Sapporo-like human caliciviruses. J Virol 73(962):5–31
Kojima S, Kageyama T, Fukushi S, Hoshino FB, Shinohara M, Uchida K, Natori K, Takeda N, Katayama K (2002) Genogroup-specific PCR primers for detection of Norwalk-like viruses. J Virol Methods 100:107–114
Guo M, Hayes J, Cho KO, Parwani AV, Lucas LM, Saif LJ (2001) Comparative pathogenesis of tissue culture-adapted and wild-type cowden porcine enteric calicivirus (PEC) in gnotobiotic pigs and induction of diarrhea by intravenous inoculation of wild-type PEC. J Virol 75(19):9239–9251
Jiang X, Huang PW, Zhong WM, Farkas T, Cubitt DW, Matson DO (1999) Design and evaluation of a primer pair that detects both Norwalk- and Sapporo-like caliciviruses by RT-PCR. J Virol Methods 83:145–154
Le Guyader F, Neill FH, Estes MK, Monroe SS, Ando T, Atmar RL (1996) Detection and analysis of a small round-structured virus strain in oysters implicated in an outbreak of acute gastroenteritis. Appl Environ Microbiol 62(11):4268–4272
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Wang QH, Souza M, Funk JA, Zhang W, Saif LJ (2006) Prevalence of noroviruses and sapoviruses in swine of various ages determined by reverse transcription-PCR and microwell hybridization assays. J Clin Microbiol 44(6):2057–2062
Mauroy A, Scipioni A, Mathijs E, Miry C, Ziant D, Thys C, Thiry E (2008) Noroviruses and sapoviruses in pigs in Belgium. Arch Virol 153:1927–1931
Wolf S, Williamson W, Hewitt J, Lin S, Rivera-Aban M, Ball A, Scholes P, Savill M, Greening GE (2009) Molecular detection of norovirus in sheep and pigs in New Zealand farms. Vet Microbiol 133:184–189
Cunha JB, de Mendonca MCL, Miagostovich MP, Leite JPG (2010) Genetic diversity of porcine enteric caliciviruses in pigs raised in Rio de Janeiro State, Brazil. Arch Virol 155:1301–1305
Sisay Z, Djikeng A, Berhe N, Belay B, Gebreyes W, Abegaz EW, Njahira NM, Wang QH, Saif LJ (2016) Prevalence and molecular characterization of human noroviruses and sapoviruses in Ethiopia. Arch Virol. doi:10.1007/s00705-016-2887-7
Chao DY, Wei JY, Chang WF, Wang J, Wang LC (2012) Detection of multiple genotypes of calicivirus infection in asymptomatic swine in Taiwan. Zoonoses Public Health 59:434–444
Farkas T, Nakajima S, Sugieda M, Deng ZW (2005) Seroprevalence of noroviruses in Swine. J Clin Microbiol 43(2):657–661
Cheetham S, Souza M, Meulia T, Grimes S, Han MG, Saif LJ (2006) Pathogenesis of a genogroup II human norovirus in gnotobiotic pigs. J Virol 80:10372–10381
Takanashi S, Hashira S, Matsunaga T, Yoshida A, Shiota T, Tung PG, Khamrin P, Okitsu S, Mizuguchi M, Igarashi T, Ushijima H (2009) Detection, genetic characterization, and quantification of norovirus RNA from sera of children with gastroenteritis. J Clin Virol 44:161–163
van Der Poel WH, Vinje J, van Der Heide R, Herrera MI, Vivo A, Koopmans MP (2000) Norwalk-like calicivirus genes in farm animals. Emerg Infect Dis 6:36–41
Acknowledgments
We thank the staff of the swine farm, particularly Dr. Nitsuh, who helped with sample collection. We are very grateful to Dr. Girmay Medhin for his assistance and encouragement. We appreciate the support of the Segolip unit of BecA-ILRI Hub in sequencing the samples from this study. We are also grateful for the cooperation and support of the Department of Molecular, Cellular and Biological Sciences, and Aklilu Lemma Institute of Pathobiology, Addis Ababa University, in facilitating the successful accomplishment of the project. We gratefully acknowledge the financial support of the African Biosciences Challenge Fund (ABCF)/ Biosciences east and central Africa (BecA). We also thank the VPH-Biotech East Africa Consortium and the National Institutes of Health-Fogarty (Fogarty Grant D43TW008650, The Ohio State University) and Dr. Wondwossen Gebreyes, for their financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This project was supported by the VPH-Biotech East Africa Consortium and the National Institutes of Health-Fogarty (Fogarty Grant D43TW008650, W Gebreyes, PI, The Ohio State University) and BecA-ILRI Hub through the Africa Biosciences Challenge Fund (ABCF) program. The ABCF Program is funded by the Australian Department for Foreign Affairs and Trade (DFAT) through the BecA-CSIRO partnership, the Syngenta Foundation for Sustainable Agriculture (SFSA), the Bill & Melinda Gates Foundation (BMGF), the UK Department for International Development (DFID), and the Swedish International Development Cooperation Agency (Sida).
Conflict of interest
None.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Rights and permissions
About this article
Cite this article
Sisay, Z., Djikeng, A., Berhe, N. et al. First detection and molecular characterization of sapoviruses and noroviruses with zoonotic potential in swine in Ethiopia. Arch Virol 161, 2739–2747 (2016). https://doi.org/10.1007/s00705-016-2974-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-016-2974-9