Abstract
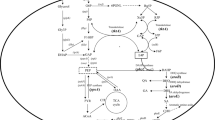
Trans-2,3-dihydro-3-hydroxyanthranilic acid (DHHA) is a cyclic β-amino acid that can be used for the synthesis of chiral materials and nonnatural peptides. The aim of this study was to accumulate DHHA by engineering Pseudomonas chlororaphis GP72, a nonpathogenic strain that produces phenazine-1-carboxylic acid and 2-hydroxyphenazine. First, the phzF deletion mutant DA1 was constructed, which produced 1.91 g/L DHHA. Moreover, rpeA and pykF were disrupted and then ppsA and tktA were co-expressed in strain DA1. The resulting strain DA4 increased DHHA concentration to 4.98 g/L, which is 2.6-fold than that of DA1. The effects of the addition of glucose, glycerol, l-tryptophan, and Fe3+on DHHA production were also investigated. Strain DA4 produced 7.48 g/L of DHHA in the culture medium in the presence of 12 g/L glucose and 3 mM Fe3+, which was 1.5-fold higher than the strain in the original fermentation conditions. These results indicate the potential of P. chlororaphis GP72 as a DHHA producer.






Similar content being viewed by others
References
Blankenfeldt W, Kuzin AP, Skarina T, Korniyenko Y, Tong L, Bayer P, Janning P, Thomashow LS, Mavrodi DV (2004) Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens. Proc Natl Acad Sci 101:16431–16436
Bongaerts J, Esser S, Lorbach V, Al-Momani L, Muller MA, Franke D, Grondal C, Kurutsch A, Bujnicki R, Takors R, Raeven L, Wubbolts M, Bovenberg R, Nieger M, Schurmann M, Trachtmann N, Kozak S, Sprenger GA, Muller M (2011) Diversity-oriented production of metabolites derived from chorismate and their use in organic synthesis. Angew Chem 123:7927–7932
Bunnage ME, Ganesh T, Masesane IB, Orton D, Steel PG (2003) Asymmetric synthesis of the putative structure of (−)-oryzoxymycin. Org Lett 5:239–242
Dosselaere F, Vanderleyden J (2001) A metabolic node in action: chorismate-utilizing enzymes in microorganisms. Crit Rev Microbiol 27:75–131
Gosset G (2009) Production of aromatic compounds in bacteria. Curr Opin Biotechnol 20:651–658
Gunther NW, Nunez A, Fett W, Solaiman DK (2005) Production of rhamnolipids by Pseudomonas chlororaphis, a nonpathogenic bacterium. Appl Environ Microbiol 71:2288–2293
Haas D, Defago G (2005) Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 3:307–319
Heckman KL, Pease LR (2007) Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2:924–932
Hoffmann A, Thimm T, Droge M, Moore ER, Munch JC, Tebbe CC (1998) Intergeneric transfer of conjugative and mobilizable plasmids harbored by Escherichia coli in the gut of the soil microarthropod Folsomia candida (Collembola). Appl Environ Microbiol 64:2652–2659
Huang L, Chen MM, Wang W, Hu HB, Peng HS, Xu YQ, Zhang XH (2011) Enhanced production of 2-hydroxyphenazine in Pseudomonas chlororaphis GP72. Appl Microbiol Biotechnol 89:169–177
Jiang M, Zhang H (2016) Engineering the shikimate pathway for biosynthesis of molecules with pharmaceutical activities in E. coli. Curr Opin Biotechnol 42:1–6
Juaristi E, Soloshonok V (2005) Enantioselective synthesis of beta-amino acids. Wiley, New York
Kanner D, Gerber N, Bartha R (1978) Pattern of phenazine pigment production by a strain of Pseudomonas aeruginosa. J Bacteriol 134:690–692
Lee TS, Krupa RA, Zhang F, Hajimorad M, Holtz WJ, Prasad N, Lee SK, Keasling JD (2011) BglBrick vectors and datasheets: a synthetic biology platform for gene expression. J Biol Eng 5:1
Liu H, He Y, Jiang H, Peng H, Zhang X, Thomashow LS, Xu Y (2007) Characterization of a phenazine-producing strain Pseudomonas chlororaphis GP72 with broad-spectrum antifungal activity from green pepper rhizosphere. Curr Microbiol 54:302–306
Liu K, Hu H, Wang W, Zhang X (2016) Genetic engineering of Pseudomonas chlororaphis GP72 for the enhanced production of 2-Hydroxyphenazine. Microb Cell Factories 15:131
Mavrodi DV, Blankenfeldt W, Thomashow LS (2006) Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu Rev Phytopathol 44:417–445
McCormick J, Reichenthal J, Hirsch U, Sjolander NO (1961) (+) trans-2, 3-dihydro-3-hydroxyanthranilic acid. A new amino acid produced by Streptomyces aureofaciens. J Am Chem Soc 83:4104–4105
McDonald M, Mavrodi DV, Thomashow LS, Floss HG (2001) Phenazine biosynthesis in Pseudomonas fluorescens: branchpoint from the primary shikimate biosynthetic pathway and role of phenazine-1,6-dicarboxylic acid. J Am Chem Soc 123:9459–9460
Meade TJ (1994) Synthesis of aromatic heterocyclic polymers from a biosynthetically prepared precursor. US Patent 5,340,913,1994
Nikel PI, Kim J, Lorenzo V (2014) Metabolic and regulatory rearrangements underlying glycerol metabolism in Pseudomonas putida KT2440. Environ Microbiol 16:239–254
Noda S, Shirai T, Oyama S, Kondo A (2016) Metabolic design of a platform Escherichia coli strain producing various chorismate derivatives. Metab Eng 33:119–129
Palko M, Kiss L, Fulop F (2005) Syntheses of hydroxylated cyclic β-amino acid derivatives. Curr Med Chem 12:3063–3083
Parsons JF, Song F, Parsons L, Calabrese K, Eisenstein E, Ladner JE (2004) Structure and function of the phenazine biosynthesis protein PhzF from Pseudomonas fluorescens 2-79. Biochemistry 43:12427–12435
Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73
Sengupta S, Jonnalagadda S, Goonewardena L, Juturu V (2015) Metabolic engineering of a novel muconic acid biosynthesis pathway via 4-hydroxybenzoic acid in Escherichia coli. Appl Environ Microbiol 81:8037–8043
Shtark OY, Shaposhnikov A, Kravchenko L (2003) The production of antifungal metabolites by Pseudomonas chlororaphis grown on different nutrient sources. Microbiology 72:574–578
Slininger PJ, Jackson MA (1992) Nutritional factors regulating growth and accumulation of phenazine 1-carboxylic acid by Pseudomonas fluorescens 2-79. Appl Microbiol Biotechnol 37:388–392
Slininger P, Shea-Wilbur M (1995) Liquid-culture pH, temperature, and carbon (not nitrogen) source regulate phenazine productivity of the take-all biocontrol agent Pseudomonas fluorescens 2-79. Appl Microbiol Biotechnol 43:794–800
Sprenger GA (2007) From scratch to value: engineering Escherichia coli wild type cells to the production of L-phenylalanine and other fine chemicals derived from chorismate. Appl Microbiol Biotechnol 75:739–749
van Rij ET, Wesselink M, Chin-A-Woeng TF, Bloemberg GV, Lugtenberg BJ (2004) Influence of environmental conditions on the production of phenazine-1-carboxamide by Pseudomonas chlororaphis PCL1391. Mol Plant-Microbe Interact 17:557–566
Wang D, Yu JM, Pierson LS, Pierson EA (2012) Differential regulation of phenazine biosynthesis by RpeA and RpeB in Pseudomonas chlororaphis 30-84. Microbiology 158:1745–1757
Whistler CA, Pierson LS III (2003) Repression of phenazine antibiotic production in Pseudomonas aureofaciens strain 30-84 by RpeA. J Bacteriol 185:3718–3725
Zhang H, Pereira B, Li Z, Stephanopoulos G (2015) Engineering Escherichia coli coculture systems for the production of biochemical products. Proc Natl Acad Sci 112:8266–8271
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 21377082).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 325 kb)
Rights and permissions
About this article
Cite this article
Hu, H., Li, Y., Liu, K. et al. Production of trans-2,3-dihydro-3-hydroxyanthranilic acid by engineered Pseudomonas chlororaphis GP72. Appl Microbiol Biotechnol 101, 6607–6613 (2017). https://doi.org/10.1007/s00253-017-8408-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8408-0