Abstract
Background
Fluoroquinolones are among the most frequently utilized antibacterial agents in developing countries like Ethiopia. Ciprofloxacin has become the most prescribed drug within this class and remains as one of the top three antibacterial agents prescribed in Ethiopia. However, several studies indicated that there is a gradual increase of antibacterial resistance. Therefore, this meta-analysis aimed to quantitatively estimate the prevalence of ciprofloxacin resistance bacterial isolates in Ethiopia.
Methods
Literature search was conducted from electronic databases and indexing services including EMBASE (Ovid interface), PubMed/MEDLINE, Google Scholar, Science Direct and WorldCat. Data were extracted with structured format prepared in Microsoft Excel and exported to STATA 15.0 software for the analyses. Pooled estimation of outcomes was performed with DerSimonian-Laird random-effects model at 95% confidence level. Degree of heterogeneity of studies was presented with I2 statistics. Publication bias was conducted with comprehensive meta-analysis version 3 software and presented with funnel plots of standard error supplemented by Begg’s and Egger’s tests. The study protocol has been registered on PROSPERO with reference number ID: CRD42018097047.
Results
A total of 37 studies were included for this study. The pooled prevalence of resistance in selected gram-positive bacterial isolates against ciprofloxacin was found to be 19.0% (95% confidence interval [CI]: 15.0, 23.0). The degree of resistance among Staphylococcus aureus, Coagulase negative Staphyloccoci (CoNS), Enterococcus faecalis and Group B Streptococci (GBS) was found to be 18.6, 21.6, 23.9, and 7.40%, respectively. The pooled prevalence of resistance in gram-negative bacteria was about 21.0% (95% CI: 17, 25). Higher estimates were observed in Neisseria gonorrhea (48.1%), Escherichia coli (24.3%) and Klebsiella pneumonia (23.2%). Subgroup analysis indicated that blood and urine were found to be a major source of resistant S. aureus isolates. Urine was also a major source of resistant strains for CoNS, Klebsiella and Proteus species.
Conclusion
Among gram-positive bacteria, high prevalence of resistance was observed in E. faecalis and CoNS whereas relatively low estimate of resistance was observed among GBS isolates. Within gram-negative bacteria, nearly half of isolates in N. gonorrhoea were found ciprofloxacin resistant. From enterobacteriaceae isolates, K. pneumonia and E. coli showed higher estimates of ciprofloxacin resistance.
Similar content being viewed by others
Background
Quinolones are groups of antibacterial drugs having an extensive application in both clinical and veterinary medicine. The older (first generation) quinolones including nalidixic acid and cinoxacin were primarily used for the treatment of urinary tract infections as their concentration in urine is relatively higher than that of the plasma. In 1980s, the introduction of fluorinated derivatives (fluoroquinolones) such as ciprofloxacin and norfloxacin became a major breakthrough in the development of relatively safer, orally effective and entirely synthetic broad spectrum antibacterial agents [1, 2]. As a result, quinolones have been routinely used for several bacterial infections. Recently, ciprofloxacin was pointed out as the most consumed antibacterial agent world-wide. Within a second generation quinolones, it has a sound medical importance in treating infections caused by many enterobacteriaceae and other gram-negative bacilli. Ciprofloxacin is the most potent of fluoroquinolones for pseudomonal infections associated with cystic fibrosis. However, their widespread use with some degree of evidence of misuse or use of these agents to micro-organisms to which they have poor activity has been blamed for the rapid development of resistance to these agents [3, 4].
In Ethiopia, ciprofloxacin has become the most commonly utilized fluoroquinolone and one of the top three antibacterial agents in clinical practice [5,6,7,8]. Study conducted by Birru et al. indicated that there is a high degree of inappropriate use of ciprofloxacin. The study emphasized that nearly half of the treatment was shown to have inappropriate dosage regimen with the duration of therapy being the dominant one in Boru Meda Hospital [9]. Such inappropriate use paves a way forward for the emergence and spread of antimicrobial resistance (AMR). AMR can result from mutations in housekeeping structural or regulatory genes as well as from horizontal acquisition of foreign genetic information [10,11,12]. Resistance to the quinolones often emerges at low-levels by acquisition of an initial resistance conferring mutation. Acquisition of subsequent mutations leads to higher levels of resistance against second and newer-generation quinolones such as ciprofloxacin [13]. At present, AMR is resulting in increased morbidity, mortality, and healthcare costs in developing countries [14]. This study is, therefore, aimed to quantitatively estimate ciprofloxacin resistance among clinically relevant bacterial isolates in Ethiopia.
Methods
Study protocol
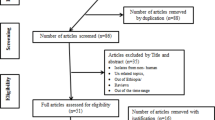
The identification of records, screening of titles and abstracts as well as evaluation of eligibility of full texts for final inclusion was conducted in accordance with the Preferred Reporting Items for Systematic review and Meta-analysis (PRISMA) flow diagram [15]. PRISMA checklist [16] was also strictly followed while conducting this systematic review and meta-analysis. The completed checklist has been provided as supplementary material (Additional file 1: Table S1). The study protocol is registered on PROSPERO with reference number ID: CRD42018097047 and the published methodology is available online from: http://www.crd.york.ac.uk/PROSPERO/display_record.php?ID=CRD42018097047
Identification of records and search strategy
Literature search was carried out through visiting legitimate databases and indexing services-PubMed/MEDLINE, EMBASE (Ovid interface) and other supplementary sources including Google Scholar, WorldCat catalog, ResearchGate and Cochrane library. Advanced search strategies were applied in major databases to retrieve relevant findings closely related to resistance/susceptibility of isolates to ciprofloxacin. Articles published in subscription based journals under Science-Direct and Wiley online library were accessed through HINARI:WHO for developing countries. The search was conducted with the aid of carefully selected key-words and indexing terms within specified time (online records from 2015- May, 2018). Excluding the non-explanatory terms, the search strategy included “quinolone [MeSH]”, ciprofloxacin [MeSH], “antimicrobial susceptibility”, “antimicrobial resistance”, “antibacterial sensitivity” and “Ethiopia”. Boolean operators (AND, OR), truncation and MeSH terms were used appropriately for systematic identification of records for the research question. The search was conducted from 25 April to 10 May, 2018 and all published and unpublished articles available online till the day of data collection were considered. Gray literatures from organizations and online university repositories were accessed through Google Scholar and WorldCat.
Screening and eligibility of studies
Records identified from various electronic databases, indexing services and directories were exported to ENDNOTE reference software version 8.2 (Thomson Reuters, Stamford, CT, USA) with compatible formats. Duplicate records were identified, recorded and removed with ENDNOTE. Some duplicates were addressed manually due to variation in reference styles across sources. Thereafter, two authors (MS and FW) independently screened the title and abstracts with predefined inclusion criteria. Two authors (MS and TT) also independently collected full texts and evaluated the eligibility of them for final inclusion. In each case, the rest authors played a critical role in solving discrepancies arose between two authors to come into consensus.
Inclusion and exclusion criteria
During initial screening of titles and abstracts as well as evaluating full texts for eligibility, there have been predefined inclusion-exclusion criteria. Cross sectional studies addressing the prevalence of ciprofloxacin-resistant bacterial isolates obtained from human source (patients) regardless of the clinical characteristics and nature of specimen were included. Only English language literatures and online records published from 2015 to May, 2018 were considered for further eligibility assessment. All review articles and original articles conducted outside of Ethiopia were excluded during initial screening. Articles with irretrievable full texts (after requesting full texts from the corresponding authors via email and/or ResearchGate), records with unrelated outcome measures, articles with missing or insufficient outcomes were excluded.
Data extraction
With the help of standardized data abstraction format prepared in Microsoft Excel (Additional file 2: Table S2), two authors (MS and HM) independently extracted important data related to study characteristics (study area, first author, year of publication, study design, patient characteristics, source of isolates, types of isolates, and number of isolates) and outcome of interest (number of resistant isolates for each bacterium).
Critical appraisal of studies
The quality of studies was evaluated according to Newcastle-Ottawa scale adapted for cross-sectional studies [17] and graded out of 10 points (stars). For ease of assessment, the tool has included important indicators categorized in to three major sections: 1) the first secstion assesses the methodological quality of each study and weighs a maximum of five stars 2) the second section considers comparability of the study and takes 2 stars 3) the remaining section assess outcomes with related to statistical analysis. This critical appraisal was conducted to assess the internal (systematic error) and external validity of studies and to reduce the risk of biases in individual studies. The mean score of two authors were taken for final decision and studies with score greater than or equal to five were included.
Outcome measurements
The primary outcome measure is the prevalence of ciprofloxacin resistant bacterial isolates in Ethiopia. It is aimed to assess the pooled estimates of antibacterial resistance at the national level. The measurement was conducted for selected gram-positive (Staphylococcous aureus; Coagulase negative staphylococci (CoNS), Group B Streptococci (GBS) and Enterococcus faecalis) and gram-negative bacterial isolates (Escherichia coli, Klebsiella pneumonia, Pseudomonas aueroginosa, Proteus species, Neisseria gonorrhea, and other enteric microorganisms) obtained from patients with presumed or confirmed infectious diseases. Subgroup analysis was also conducted based on the source of bacterial isolates.
Data processing and statistical analysis
The relevant data were extracted from included studies using format prepared in Microsoft Excel and exported to STATA 15.0 for outcome measures and subgroup analyses. Considering variation in true effect sizes across population (clinical heterogeneity), Der Simonian and Laird’s random effects model was applied for the analyses at 95% confidence level. Heterogeneity of studies was determined using I2 statistics. Comprehensive Meta-analysis version-3 software (Biostat, Englewood, New Jersey, USA) was used for publication bias assessment. For gram-positive and gram-negative bacterial isolates, the presence of publication bias was evaluated by using the Begg’s and Egger’s tests and presented with funnel plots of standard error of Logit event rate [18, 19]. A statistical test with a p-value less than 0.05 (one tailed) was considered significant.
Results
Search results
A total of 416 records were identified from several sources including PubMed/MEDLINE, EMBASE, Google Scholar, Science Direct and WorldCat catalog. From these, 137 duplicate articles were removed with the help of ENDNOTE and manual tracing. The remaining 279 records were screened using their titles and abstracts and 225 of them were excluded. Full texts of 54 records were then evaluated for eligibility. From these, 17 articles were also excluded as the outcome of interest was found missing, insufficient and/or ambiguous. Finally, 37 articles have passed the eligibility criteria and quality assessment and hence included in the study (Fig. 1).
Study characteristics
As shown in Table 1, a total of 37 studies with 3235 selected bacterial isolates (1303 gram-positive and 1932 gram-negative) were included for systematic review and meta-analysis. We included studies that employed both retrospective and prospective cross-sectional study design. The year of publication of included studies ranged from 2015 to 10 May 2018 since antimicrobial resistance is highly time-sensitive. The study included a wide range of clinical characteristics of patients, sources of isolates (specimens), nature of bacterial isolates and effect sizes. Patients with presumed or confirmed urinary tract infections took larger proportion of participants and midstream urine sample was the major source of bacterial isolates [20,21,22,23,24,25,26,27,28,29]. The rest sources of isolates were blood from septicemia and febrile patients [30,31,32,33,34], stool from patients with acute diarrhea [35,36,37,38,39], external ocular discharges from patients with ocular infections [40,41,42], vaginal discharges from pregnant women with infections [43,44,45], ear discharges with bacterial otitis media [46,47,48], and wound swabs from infected wounds [49, 50], among others. Some samples were taken from more than one source in a given patient [51,52,53,54,55]. Six of the included studies were retrospective analyses of secondary data [23, 28, 46, 47, 49, 52]. Majority of the isolates from stool were enteric gram-negative micro-organisms (salmonella and shigella) and gram positive enterococci. The average quality scores of studies ranged from 6 to 10 as per the Newcastle-Ottawa scale (Table 1).
Study outcome measures
Gram-positive bacteria
The overall estimate of resistance in selected gram-positive bacterial isolates against ciprofloxacin was found to be 19% (95% CI: 15, 23) (Fig. 2). In this category, the pooled estimates of resistance in S. auerus was 18.6% (95% CI: 13.5, 23.7) with degree of heterogeneity (I2), 88.18%. The resistance level of CoNS isolates was found to be 21.6% (95% CI: 12.4, 30.8). Higher degree of resistance was observed among Enterococcus faecalis with prevalence rate of 23.9%. There was low level of ciprofloxacin resistance in GBS isolates (7.40%) (Table 2).
Gram-negative bacteria
The gram-negative bacteria were the most common isolates obtained from several sources. The pooled estimate of resistance was 21% (95% CI: 17, 25) (Fig. 3). Among the selected isolates, higher degree of resistance was observed in N. gonorrhea, E. coli and K. pneumoniae with prevalence of 48.1, 24.3 and 23.2%, respectively. Besides, the pooled estimates of resistance in Proteus species (mainly P. mirabilis) and Pseudomonas species (primarily P. aueroginosa) against ciprofloxacin were found to be 16.0% (95% CI: 7.9, 24.1) and 14.1% (95% CI: 8.3, 19.8), respectively. The lowest degree of resistance was found among other gram negative enteric pathogens (salmonella and shigella) obtained from stool in patients with acute diarrhea. The overall estimate of resistance in these enteric species was found to be 6.3% (95% CI: 1.5, 11.1). Individual isolate (subgroup analysis) indicated that the prevalence of resistance in salmonella and shigella species was 8.1 and 5.8%, respectively (Table 2). In addition, we performed a univariate meta-regression model to identify whether sample size of individual isolates is a possible sources of heterogeneity; however, only the sampling distribution of S. aureus was found to be statistically significant (p value = 0.005) (Table 3).
Source based subgroup and sensitivity analyses
There was a significant change on the degree of heterogeneity when we had excluded the expected outliers and studies with few numbers of isolates (less than five) per bacterium from the analyses. Very few sample size significantly affected the confidence intervals and point estimates. Therefore, we were subjected to exclude some of the studies for the meta-analysis at the initial scenario. We also conducted a subgroup analysis based on the source of bacterial isolates. These analyses further clarify whether there is a clinically significant difference in the degree of resistance of bacterial isolates across sources of specimens. Highest prevalence of resistant isolates was obtained from urine sample for CoNS (36%), K. pneumoniae (32%) and Proteus species (40%). Among the common sources, blood sample was endowed with larger proportion of resistant isolates of S. aureus 33% (95% CI: 20, 45). The resistance rates of E. coli and Pseudomonas spp from wound swabs and vaginal discharges, respectively, was found to be high (Table 4).
Publication bias
Funnel plots of standard error with Logit event rate (proportion of resistant isolates) supplemented by statistical tests confirmed that there is some evidence of publication bias on studies reporting the prevalence of ciprofloxacin resistance among gram-positive (Begg’s test, p = 0.086; Egger’s test, p = 0.026) and gram- negative bacteria (Begg’s test, p = 0.06; Egger’s test, p = 0.0003) (Fig. 4a and b).
Discussion
This systematic review and meta-analysis included 37 original studies addressing the prevalence of ciprofloxacin-resistant clinical isolates in Ethiopia within the specified timeframe. Regardless of the source and identity of isolates, the study revealed that one in five clinical isolates were found to be ciprofloxacin resistant in both gram-positive and gram-negative bacteria. E. faecalis from gram-positive bacteria and N. gonorrhoea, E. coli and K. pneumoniae from gram-negative bacteria exhibited higher prevalence of resistance as the meta-analysis indicated. The resistance estimate in other enteric pathogens (shigella and salmonella), obtained from stool samples, were found to be relatively less in Ethiopia. Urine and blood samples have been the major source of resistant isolates. In spite of relatively low level of resistance (8.1%) in Ethiopia, the emergence of ciprofloxacin resistance in common salmonella serotypes worldwide is becoming a serious public health concern. Besides, resistance to the first generation quinolones (nalidixic acid) has been associated with reduced efficacy of 6-fluorinated-quinolones such as ciprofloxacin [56, 57].
Routine antimicrobial surveillance data indicated the presence of strong relationship between antimicrobial use and resistance at a national level in Europe [58]. Even if quinolones are less likely to select for resistance compared to other natural antibiotics, highly level of use with some degree of misuse facilitates resistance selection and spread of quinolones resistance (QNR) genes to areas where the prevalence of resistance is found to be low [59,60,61,62,63,64]. Population mobility is a main factor in the spread of antimicrobial–resistant organisms [64]. To this end, Vernet et al. reported that 65% of E. coli strains isolated from patients who had traveled to India were found resistant to quinolones including ciprofloxacin [65]. Besides, surveillance data showed that resistance in E. coli and K. pneumonia has become consistently higher for antimicrobial agents that have been in use for long time in human and veterinary medicine [12]. In trajectory with our findings, significant increment in resistant level of K. pneumonia strain against ciprofloxacin was observed from 1998 to 2010 in United States [66]. Even if fluoroquinolones such as ciprofloxacin and ofloxacin have been highly effective in treating gonorrhea, the widespread and often inappropriate use leads to the emergence of fluoroquinolone resistant N. gonorrhoea [4, 67]. World Health Organization updated the current treatment profiles of N. gonorrhea as there has been an established resistance reports from various regions [67, 68].
To date, several mechanisms of quinolone resistance have been determined: modification of bacterial targets (DNA gyrase or topoisomerase IV) to which quinolones bind, decreased intracellular (bacterial) concentration due to an over-expression of active efflux pumps and enzymatic inactivation (acetylation) of quinolones, among others. Recently, mobile genetic elements carrying the QNR gene, which confer resistance to quinolones, have also been described [1, 69,70,71]. Amino acid changes in critical regions of the enzyme-DNA complex (quinolone resistance–determining region [QRDR]) reduce quinolone affinity for both targets [59,60,61]. QRDR mutation was identified in Enterococcus isolates; with serine being changed in gyrA83, gyrA87 and parC80. This result showed that gyrA and parC mutations could be important factors for high-level of resistance to such species against ciprofloxacin [70]. QNR proteins protect target enzymes from quinolone inhibition. The AAC(6′)-Ib-cr determinant acetylates several fluoroquinolones, such as norfloxacin and ciprofloxacin [69].
Plasmid-mediated quinolone resistance has been shown to compromise the bactericidal activity of fluoroquinolones when expressed in Enterobacteriaceae [72]. For example, plasmidic transfer of genes has resulted in spread of resistant strains among E. coli, K. pneumoniae, and Proteus species [73]. Jacoby et al. described the presence of QNR gene up on analyzing a long series of gram-negative microorganisms (mainly K. pneumonia and E. coli) from different geographical origins (19 countries) around the world) [62]. The development of fluoroquinolone resistance in staphylococci, P. aeruginosa, and other pathogens can also occur through alterations in DNA topoisomerase [74]. Besides, an endogenous system which actively transports quinolones out of the bacteria was described initially in E. coli and later in other gram-negative and gram-positive bacteria such as S. aureus [75, 76]. The QepA and OqxAB efflux pumps extrude fluoroquinolones from the bacterial cell [69]. Generally, the above-mentioned mechanisms of resistance have been established upon routine exposure of quinolones for treatment of many bacterial infections. AMR has resulted in increased morbidity, mortality, as well as direct and indirect healthcare costs in developing countries [14]. A notable example is an epidemic of infection associated with ciprofloxacin resistant S. typhi observed in Tajikistan [77].
Conclusion
The study revealed that one in five gram-positive or gram-negative bacterial isolates developed resistance against ciprofloxacin in Ethiopia. Among gram-positive bacteria, high level of resistance was observed in Enterococci and CoNS whereas and relatively low degree of resistance was observed among GBS isolates. Within gram-negative bacteria, nearly half of isolates of N. gonorrhoeae was found ciprofloxacin resistant. From enterobacteriaceae isolates, K. pneumonia and E. coli showed relatively higher degree of ciprofloxacin resistance while shigella and salmonella had low level of resistance. Urine and Blood samples were the major sources of ciprofloxacin resistant isolates. Considering resistance estimates in to account, antimicrobial stewardship programs should be established in Ethiopian healthcare settings thereby preserves antimicrobials and contains AMR.
Abbreviations
- AMR:
-
Antimicrobial resistance
- CoNS:
-
Coagulase negative Staphylococci
- GBS:
-
Group B Streptococci
- MeSH:
-
Medical subject headings
- PRISMA:
-
Preferred Reporting Items for Systematic Review and Meta-analysis
- QNR:
-
Quinolone resistance
- QRDR:
-
Quinolone resistance determining region
- WHO:
-
World Health Organization
References
Ruiz J. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother. 2003;51(5):1109–17.
WHO. World medicine situation report 2011. 2016.
Solomon DH, Van Houten L, Glynn RJ, Baden L, Curtis K, Schrager H, Avorn J. Academic detailing to improve use of broad-spectrum antibiotics at an academic medical center. Arch Intern Med. 2001;161(15):1897–902.
Hart C, Kariuki S. Antimicrobial resistance in developing countries. BMJ. 1998;317(7159):647.
Jabo SA, Tebeka AG, Asebe DS, Beri FZ, Temesgen DM, Beyene AT, Gemeda HB, Genemo AA, Arega ED, Yilma FN. Assessment of medication prescription pattern at bole health center, Ethiopia. Int J Sci Rep. 2017;4(1):15–8.
Sisay M, Mengistu G, Molla B, Amare F, Gabriel T. Evaluation of rational drug use based on World Health Organization core drug use indicators in selected public hospitals of eastern Ethiopia: a cross sectional study. BMC Health Serv Res. 2017;17(1):161.
Berha AB, Seyoum N. Evaluation of drug prescription pattern using World Health Organization prescribing indicators in Tikur Anbessa specialized hospital: a cross–sectional study. J Drug Deliv Therapeut. 2018;8(1):74–80.
Tulu S, Tadesse T, Alemayehu Gube A. Assessment of antibiotic utilization pattern in treatment of acute Diarrhoea diseases in Bishoftu general hospital, Oromia Ethiopia. Adv Med. 2018;2018:2376825.
Biru TT, Defersha AD, Gelaw BK, Tegegne GT. Drug utilization review of ciprofloxacin in the outpatient department of boru Meda hospital, south wollo zone, Amhara region, Ethiopia. Int J Basic Clin Pharmacol. 2017;3(1):171–8.
Courvalin P. Antimicrobial drug resistance: “prediction is very difficult, especially about the future”. Emerg Infect Dis. 2005;11(10):1503.
WHO. Antimicrobial resistance: global report on surveillance: World Health Organization; 2014. Available at: http://www.who.int/drugresistance/documents/surveillancereport/en/. Accessed 15 Aug 2018.
Tadesse DA, Zhao S, Tong E, Ayers S, Singh A, Bartholomew MJ, McDermott PF. Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950–2002. Emerg Infect Dis. 2012;18(5):741.
Morgan-Linnell SK, Zechiedrich L. Contributions of the combined effects of topoisomerase mutations toward fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 2007;51(11):4205–8.
Cohen ML. Epidemiology of drug resistance: implications for a post—antimicrobial era. Science. 1992;257(5073):1050–5.
Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097.
Moher D, Liberati A, Tetzlaff J, Altman D: Prisma 2009 checklist available at: www prisma-statement org. Accessed 28 Apr 2018. 2009.
Newcastle- Ottawa: Scale customized for cross-sectional studies In. available from: https://static-contentspringer.com/esm//12889_2012_5111_MOESM3_ESMdoc. Accessed 27 July 2018.
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–101.
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–34.
Ayelign B, Abebe B, Shibeshi A, Meshesha S, Shibabaw T, Addis Z, Gelaw A, Dagnew M. Bacterial isolates and their antimicrobial susceptibility patterns among pediatric patients with urinary tract infections. Turk J Urol. 2018;44(1):62–9.
Bekele T, Tesfaye A, Sewunet T, Waktola HD. Pseudomonas aeruginosa isolates and their antimicrobial susceptibility pattern among catheterized patients at Jimma University teaching hospital, Jimma Ethiopia. BMC Res Notes. 2015;8:488.
Bitew A, Molalign T, Chanie M. Species distribution and antibiotic susceptibility profile of bacterial uropathogens among patients complaining urinary tract infections. BMC Infect Dis. 2017;17(1) (no pagination):654.
Derbie A, Hailu D, Mekonnen D, Abera B, Yitayew G. Antibiogram profile of uropathogens isolated at Bahir Dar regional Health Research Laboratory Centre, Northwest Ethiopia. Pan Afr Med J. 2017;26:134.
Dereje M, Woldeamanuel Y, Asrat D, Ayenachew F. Urinary tract infection among fistula patients admitted at Hamlin fistula hospital, Addis Ababa, Ethiopia. BMC Infect Dis. 2017;17(1):150.
Derese B, Kedir H, Teklemariam Z, Weldegebreal F, Balakrishnan S. Bacterial profile of urinary tract infection and antimicrobial susceptibility pattern among pregnant women attending at antenatal Clinic in Dil Chora Referral Hospital, Dire Dawa, eastern Ethiopia. Ther Clin Risk Manag. 2016;12:251–60.
Eshetie S, Unakal C, Gelaw A, Ayelign B, Endris M, Moges F. Multidrug resistant and carbapenemase producing Enterobacteriaceae among patients with urinary tract infection at referral hospital, Northwest Ethiopia. Antimicrob Resist Infect Control. 2015;4:12.
Gezmu T, Regassa B, Manilal A, Mama M, Merdekios B. Prevalence, diversity and antimicrobial resistance of bacteria isolated from the UTI patients of Arba Minch Province, southern Ethiopia. Transl Biomed. 2016;7:3.
Mitiku SY. Bacterial uropathogens and their antibiotic susceptibility pattern at Dessie regional health laboratory. J Microbiol Biotechnol Res. 2017;3:1–9.
Nigussie D, Amsalu A. Prevalence of uropathogen and their antibiotic resistance pattern among diabetic patients. Turk J Urol. 2017;43(1):85–92.
Kumalo A, Kassa T, Daka D, Tadesse AH. Bacterial profile of adult sepsis and their antimicrobial susceptibility pattern at Jimma University specialized hospital, south West Ethiopia. Health Sci J. 2016;10:2.
Negussie A, Mulugeta G, Bedru A, Ali I, Shimeles D, Lema T, Aseffa A. Bacteriological profile and antimicrobial susceptibility pattern of blood culture isolates among septicemia suspected children in selected hospitals Addis Ababa, Ethiopia. Int J Biol Med Res. 2015;6(1):4709.
Wasihun AG, Wlekidan LN, Gebremariam SA, Dejene TA, Welderufael AL, Haile TD, Muthupandian S. Bacteriological profile and antimicrobial susceptibility patterns of blood culture isolates among febrile patients in Mekelle hospital, northern Ethiopia. Springerplus. 2015;4:314.
Wasihun AG, Wlekidan LN, Gebremariam SA, Welderufael AL, Muthupandian S, Haile TD, Dejene TA. Diagnosis and treatment of typhoid fever and associated prevailing drug resistance in northern Ethiopia. Int J Infect Dis. 2015;35:96–102.
Hailu D, Abera B, Yitayew G, Mekonnen D, Derbie A. Bacterial blood stream infections and antibiogram among febrile patients at Bahir Dar regional Health Research Laboratory center, Ethiopia. Ethiopian J Sci Technol. 2016;9(2):103–12.
Abamecha A, Wondafrash B, Abdissa A. Antimicrobial resistance profile of enterococcus species isolated from intestinal tracts of hospitalized patients in Jimma, Ethiopia. BMC Res Notes. 2015;8:213.
Ameya G, Tsalla T, Getu F, Getu E. Antimicrobial susceptibility pattern, and associated factors of salmonella and Shigella infections among under five children in Arba Minch, South Ethiopia. Ann Clin Microbiol Antimicrob. 2018;17(1) (no pagination):1.
Gebrekidan A, Dejene TA, Kahsay G, Wasihun AG. Prevalence and antimicrobial susceptibility patterns of Shigella among acute diarrheal outpatients in Mekelle hospital, northern Ethiopia. BMC Res Notes. 2015;8:611.
Lamboro T, Ketema T, Bacha K. Prevalence and antimicrobial resistance in salmonella and Shigella species isolated from outpatients, Jimma University specialized hospital, Southwest Ethiopia. Can J Infect Dis Med Microbiol. 2016;2016:4210760.
Terfassa A, Jida M. Prevalence and antibiotics susceptibility pattern of salmonella and Shigella species among diarrheal patients attending Nekemte referral hospital, Oromia. Ethiopia. 2018;2018:9214689.
Teweldemedhin M, Saravanan M. Ocular bacterial infections at Quiha ophthalmic hospital, northern Ethiopia: an evaluation according to the risk factors and the antimicrobial susceptibility of bacterial isolates. BMC Infect Dis. 2017;17(1):207.
Getahun E, Gelaw B, Assefa A, Assefa Y, Amsalu A. Bacterial pathogens associated with external ocular infections alongside eminent proportion of multidrug resistant isolates at the University of Gondar Hospital, Northwest Ethiopia. BMC Ophthalmol. 2017;17(1):151.
Shiferaw B, Gelaw B, Assefa A, Assefa Y, Addis Z. Bacterial isolates and their antimicrobial susceptibility pattern among patients with external ocular infections at Borumeda hospital, Northeast Ethiopia. BMC Ophthalmol. 2015;15:103.
Alemseged G, Niguse S, Hailekiros H, Abdulkadir M, Saravanan M, Asmelash T. Isolation and anti-microbial susceptibility pattern of group B streptococcus among pregnant women attending antenatal clinics in Ayder referral hospital and Mekelle health center, Mekelle, northern Ethiopia. BMC Res Notes. 2015;8:518.
Mengist A, Kannan H, Abdissa A. Prevalence and antimicrobial susceptibility pattern of anorectal and vaginal group B streptococci isolates among pregnant women in Jimma, Ethiopia. BMC Res Notes. 2016;9:351.
Mulu W, Yimer M, Zenebe Y, Abera B. Common causes of vaginal infections and antibiotic susceptibility of aerobic bacterial isolates in women of reproductive age attending at Felegehiwot referral hospital, Ethiopia: a cross sectional study. Springerplus. 2015;15:42.
Argaw-Denboba A, Abejew AA, Mekonnen AG. Antibiotic-resistant bacteria are major threats of otitis Media in Wollo Area Northeastern Ethiopia: a ten-year retrospective analysis. Int J Microbiol. 2016;2016:8724671.
Hailu D, Mekonnen D, Derbie A, Mulu W, Abera B. Pathogenic bacteria profile and antimicrobial susceptibility patterns of ear infection at Bahir Dar regional Health Research Laboratory center, Ethiopia. Springerplus. 2016;5:466.
Wasihun AG, Zemene Y. Bacterial profile and antimicrobial susceptibility patterns of otitis media in Ayder teaching and referral hospital, Mekelle University northern Ethiopia. Springerplus. 2015;4:701.
Hailu D, Derbie A, Mekonnen D, Zenebe Y, Adem Y, Worku S, Biadglegne F. Drug resistance patterns of bacterial isolates from infected wounds at Bahir Dar regional Health Research Laboratory center, Northwest Ethiopia. Ethiop J Health Dev. 2016;30(3):112–7.
Dessie W, Mulugeta G, Fentaw S, Mihret A, Hassen M, Abebe E. Pattern of bacterial pathogens and their susceptibility isolated from surgical site infections at selected referral hospitals, Addis Ababa, Ethiopia. Int J Microbiol. 2016;2016:2418902.
Abera B, Kibret M, Mulu W. Extended-Spectrum beta (beta)-lactamases and Antibiogram in Enterobacteriaceae from clinical and drinking water Sources from Bahir Dar City, Ethiopia. PLoS One. 2016;11(11):e0166519.
Mulu W, Abera B, Yimer M, Hailu T, Ayele H, Abate D. Bacterial agents and antibiotic resistance profiles of infections from different sites that occurred among patients at Debre Markos referral hospital, Ethiopia: a cross-sectional study. BMC Res Notes. 2017;10(1):254.
Regasa B, Yilma D, Sewunet T, Beyene G. Antimicrobial susceptibility pattern of bacterial isolates from community-acquired pneumonia patients in Jimma University specialized hospital, Jimma, Ethiopia. Saudi J Health Sci. 2015;4(1):59.
Sahile T, Esseye S, Beyene G, Ali S. Post-surgical infection and antibiotic susceptibility patterns of bacteria isolated from admitted patients with signs of infection at Jimma University specialized hospital, Jimma, Ethiopia. Int J Trop Dis Health. 2016;17(4):1–12.
Assefa Y, Moges F, Endris M, Zereay B, Amare B, Bekele D, Tesfaye S, Mulu A, Belyhun Y. Bacteriological profile and drug susceptibility patterns in dacryocystitis patients attending Gondar University teaching hospital, Northwest Ethiopia. BMC Ophthalmol. 2015;15:34.
Bager F, Helmuth R. Epidemiology of resistance to quinolones in salmonella. Vet Res. 2001;32(3–4):285–90.
Mølbak K, Gerner-Smidt P, Wegener HC. Increasing quinolone resistance in salmonella enterica serotype Enteritidis. Emerg Infect Dis. 2002;8(5):514.
Van De Sande-Bruinsma N, Grundmann H, Verloo D, Tiemersma E, Monen J, Goossens H, Ferech M, System EARS: Antimicrobial drug use and resistance in Europe. Emerg Infect Dis 2008, 14(11):1722.
Lowy FD. Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest. 2003;111(9):1265–73.
Hooper DC. Fluoroquinolone resistance among gram-positive cocci. Lancet Infect Dis. 2002;2(9):530–8.
Høiby N, Jarløv JO, Kemp M, Tvede M, Bangsborg JM, Kjerulf A, Pers C, Hansen H. Excretion of ciprofloxacin in sweat and multiresistant Staphylococcus epidermidis. Lancet. 1997;349(9046):167–9.
Jacoby GA, Chow N, Waites KB. Prevalence of plasmid-mediated quinolone resistance. Antimicrob Agents Chemother. 2003;47(2):559–62.
Veldman K, Cavaco LM, Mevius D, Battisti A, Franco A, Botteldoorn N, Bruneau M, Perrin-Guyomard A, Cerny T, De Frutos Escobar C. International collaborative study on the occurrence of plasmid-mediated quinolone resistance in salmonella enterica and Escherichia coli isolated from animals, humans, food and the environment in 13 European countries. J Antimicrob Chemother. 2011;66(6):1278–86.
MacPherson DW, Gushulak BD, Baine WB, Bala S, Gubbins PO, Holtom P, Segarra-Newnham M. Population mobility, globalization, and antimicrobial drug resistance. Emerg Infect Dis. 2009;15(11):1727.
Vernet G, Mary C, Altmann DM, Doumbo O, Morpeth S, Bhutta ZA, Klugman KP. Surveillance for antimicrobial drug resistance in under-resourced countries. Emerg Infect Dis. 2014;20(3):434.
Sanchez GV, Master RN, Clark RB, Fyyaz M, Duvvuri P, Ekta G, Bordon J. Klebsiella pneumoniae antimicrobial drug resistance, United States, 1998–2010. Emerg Infect Dis. 2013;19(1):133.
CIDRAP. WHO releases its first report on global antibiotic resistance. CIDRAP. 2018. Available from: http://www.cidrap.umn.edu/news-perspective/2018/01/who-releases-its-first-report-global-antibiotic-resistance. Accessed 15 Aug 2018.
Jacoby GA. Plasmid-mediated quinolone resistance. In: Antimicrobial drug resistance: Springer; 2017. p. 265–8.
Poirel L, Cattoir V, Nordmann P. Plasmid-mediated quinolone resistance; interactions between human, animal, and environmental ecologies. Front Microbiol. 2012;3:24.
Chan M, Jo G. Characterization of antimicrobial resistance and quinolone resistance factors in highâ level ciprofloxacinâ resistant enterococcus faecalis and enterococcus faecium isolates obtained from fresh produce and fecal samples of patients. J Sci Food Agric. 2017;97:2858–64.
Wolfson JS, Hooper DC. The fluoroquinolones: structures, mechanisms of action and resistance, and spectra of activity in vitro. Antimicrob Agents Chemother. 1985;28(4):581.
Margaritis A, Galani I, Chatzikonstantinou M, Petrikkos G, Souli M. Plasmid-mediated quinolone resistance determinants among gram-negative bacteraemia isolates: a hidden threat. J Med Microbiol. 2017;66(3):266–75.
Miyakis S, Pefanis A, Tsakris A. The challenges of antimicrobial drug resistance in Greece. Clin Infect Dis. 2011;53(2):177–84.
Gold HS, Moellering RC Jr. Antimicrobial-drug resistance. N Engl J Med. 1996;335(19):1445–53.
Levy SB. Active efflux mechanisms for antimicrobial resistance. Antimicrob Agents Chemother. 1992;36(4):695.
Celesk RA, Robillard NJ. Factors influencing the accumulation of ciprofloxacin in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1989;33(11):1921–6.
Murdoch DA, Banatvaia N, Bone A, Shoismatulloev BI, Ward LR, Threlfall EJ. Epidemic ciprofloxacin-resistant salmonella typhi in Tajikistan. Lancet (London, England). 1998;351(9099):339.
Ali S, Sewunet T, Sahlemariam Z, Kibru G. Neisseria gonorrhoeae among suspects of sexually transmitted infection in Gambella hospital, Ethiopia: risk factors and drug resistance. BMC Res Notes. 2016;9(1):439.
Acknowledgments
Authors thank School of Pharmacy, and Medical Laboratory Science staffs who technically supported us for the realization of this article.
Funding
None.
Availability of data and materials
All the data is contained within the manuscript and additional files.
Author information
Authors and Affiliations
Contributions
MS, FW and TT conceived and designed the study. All authors collected scientific literatures, critically appraised individual articles for inclusion, analyzed and interpreted the findings. MS drafted the manuscript, critically reviewed it and prepared the final version for publication. All authors read and approved the final version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. Completed PRISMA checklist. The checklist highlights the important components addressed while conducting systematic review and meta-analysis from observational studies. (DOC 65 kb)
Additional file 2:
Table S2. Data abstraction format. The table presented the ways of data collection (study characteristics and outcome measures) in Microsoft excel format. It also contained a raw data for outcome analyses. (XLSX 27 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Sisay, M., Weldegebreal, F., Tesfa, T. et al. Resistance profile of clinically relevant bacterial isolates against fluoroquinolone in Ethiopia: a systematic review and meta-analysis. BMC Pharmacol Toxicol 19, 86 (2018). https://doi.org/10.1186/s40360-018-0274-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40360-018-0274-6