Abstract
Background
The burden of Klebsiella drug resistance to antimicrobials is a major public health concern worldwide; particularly the problem is severe in developing countries including Ethiopia. Therefore, the aim of this systematic review and meta-analysis is to establish the pooled estimate of Klebsiella drug resistance; and antimicrobial-specific resistance pattern among Klebsiella clinical isoaltes in Ethiopia.
Methods
Articles were searched from PubMed, Google Scholar, and Science direct and grey literature from 2009 to 2019. Four authors have independently extracted data on the prevalence and antimicrobial resistance pattern of the isolates. Statistical analysis was conducted by using Open meta-analyst (version 3.13) and Comprehensive meta-analysis (version 3.3). The main outcome measures were the overall Klebsiella resistance; and drug-specific resistance patterns. A random-effects model was used to determine the pooled resistance prevalence with 95% confidence interval (CI), and significant heterogeneity was considered at p < 0.1; and I2 > 50% using DerSimonian and Laird method. In addition, subgroup analyses were conducted to improve the outcome.
Result
We obtained 174 potentially relevant studies through searching electronic databases, and finally, 35 eligible studies were included for meta-analysis. A total of 13,269 study samples participated, from which 1017 Klebsiella species were isolated. The overall Klebsiella resistance in Ethiopia was found to stand at 53.75% (95% CI: 48.35—58.94%). Based on the subgroup analyses; the highest (64.39%); and lowest (46.16%) values were seen in Southern Nations, Nationalities, and Peoples of Ethiopia; and Tigray regions respectively; and the highest Klebsiella resistance was reported to ampicillin (90.56%), followed by amoxicillin (76.01%) and trimethoprim-sulfamethoxazole (66.91%). A relatively low level of resistance rate was observed to amikacin (16.74%) and cefoxitin (29.73%).
Conclusion
The pooled Klebsiella resistance was found to be considerably high (53.75%) to most of the essential antibiotics in Ethiopia. Klebsiella was highly resistant to ampicillin and amoxicillin but relatively lower to amikacin. Therefore, appropriate interventional strategies need to be taken to address the emerging resistance of Klebsiella species.
Similar content being viewed by others
Background
Klebsiella species are aerobic or facultative anaerobic gram negative bacteria which belong to the family Enterobacteriaceae. It is known to produce plasmid mediated extended spectrum beta lactamases (ESBLs) which breaks down antibiotics into inactive form [1] and the most common isolated species include Klebsiella pneumoniae, Klebsiella oxytoca, Klebsiella ozaenae and Klebsiella rhinoscleromatis [2, 3]. From the various species, Klebsiella pneumoniae is the most common and ubiquitous in nature causing various human infections [4, 5].
The widespread use of antimicrobials in clinical practice has led to the emergence of resistant bacterial pathogens contributing to the increased morbidity and mortality observed worldwide [6, 7]. Resistant Klebsiella is one of the opportunistic pathogen showing frequent acquisition of resistance to antibiotics accounting to about one-third of all Gram-negative infectious diseases [8, 9] such as bloodstream infection, pneumonia, urinary tract infections (UTI), nosocomial and community acquired infections [2, 10]. As there is inappropriate use of antimicrobials, resistance is tremendously increasing and the therapeutic option has been significantly reduced [11, 12]. Ultimately, this increases cost of treatment and impedes the effective prevention and treatment outcomes in clinical settings [13,14,15,16].
Nowadays, studies show that Klebsiella has been resistant to most common antibiotics including cephalosporins, monobactams, fluoroquinolones and aminoglycosides [3, 17,18,19]. This is because of the frequent empirical use of antibiotics; and persistent exposure of Klebsiella to a number of antimicrobial agents which facilitate the emergence of drug-resistant strains [20]. Previous studies have indicated Klebsiella’s resistance to antibiotics reaching 68.3% in south Africa [12], 54% in India [1], and 97.17% in Equatorial Guinea [21]. In sub-Saharan Africa including Ethiopia, the anti-microbial drug resistance is a serious problem [22]. Individual studies conducted in Ethiopia showed that the prevalence of antimicrobial resistance is high and pooled prevalence of resistance in gram negative bacteria Klebsiella pneumonia is reported to be 23.2% [23]; Klebsiella species are able to develop cross resistance; and the treatment failure is also high [3]. Moreover, one study indicates that antimicrobial resistance level of the gram-negative bacteria ranges from 20 to 100% [24]. Studies in some Ethiopian hospitals such as Jimma (77.8%) [25], Yekatit 12 (64.7%) [26] and Gondar hospitals (95.6%) [27], show that the antimicrobial resistance of Klebsiella is high. However, the comprhenssive analysis of antimicrobial drug resistance pattern of Klebsiella isolates from different parts of Ethiopia has not yet been performed nationally. Hence, the present systematic review and meta analysis was aimed at establishing the pooled prevalence of Klebsiella resistamce; and antimicrobial-specific resistance pattern among Klebsiella clinical isolates in Ethiopia.
Methods
This meta-analysis was carried out in a similar approach to the previously published studies [13, 28] and the content of this systematic review and meta-analysis is well described according to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) flow diagram and checklist [29,30,31] (S1 file).
Study selection
Exhaustive literature search was done systematically in PubMed, and Science direct, Google scholar; and manual search from Google of potentially relevant articles. The search was done by four authors (LG, TT, GGK and KBT) independently; and the search strategy was built by combining the three main arms (Table 1): Klebsiella, Antimicrobial related terms, and Ethiopia. From the citation extracted, abstracts were scanned to retrieve the clinical studies on Klebsiella infection. Studies that were relevant, by title and abstract, were accessed in full text to determine articles to be included in our meta-analysis. Finally, the references cited by each eligible study were scrutinized to identify additional source; and references were cited using endnote citation manager software (version X7)[Niles software, Clarivate analytic company]. Before starting the data extraction, the protocol was developed and we went directly to data extraction without considering the registration, because at that time there was internet interruption because of the conflict here in Tigray, Ethiopia.
Inclusion and exclusion criteria
Studies included in this comprehensive meta-analysis were those that had extractable data on drug resistance of Klebsiella isolates taken from human samples in Ethiopian health facilities or research centers. Articles published from January 2009 to December 2019 in English language; and primary researches with cross-sectional study designs were included; but review papers and other studies that were not relevant to the outcomes of interest and those that were not based on Ethiopian setting were excluded. Initially, non- relevant studies were excluded based on their titles and abstracts. From the screened papers, the duplicates, incomplete data, and studies with very small number of isolates and tested antimicrobials (less than 5 isolates and tested drugs) were excluded.
Outcome of interest
The primary outcome of interest was the prevalence of antimicrobial resistance of Klebsiella species among the total Klebsiella clinical isolates in which the prevalence was calculated by dividing the numbers of resistant Klebsiella isolates by the total number of clinically isolated Klebsiella. We also calculated the pooled resistance pattern of Klebsiella isolates to specific antibiotics as a secondary outcome of interest.
Data extraction
The process of screening (by title, abstract, and full text and data extraction) was done independently by three authors (LG, GGK, and TT), and at each step KBT was involved in consensus creation in cases of discrepancies among the three authors. During screening by title and abstract, the authors reviewed the full text of the article if needed. Data from eligible studies were extracted and summarized into an excel spreadsheet. For each of the included studies, the following information was extracted; name of regions, study design, study area/city, study names, study period, study design, types of specimens, study population, number of study participants, total numbers of isolated Klebsiella species, the average percentage of resistant Klebsiella species, antimicrobial resistance rate of Klebsiella species and references. As all of the articles used in this study are cross-sectional, the score for the quality of the study was assessed using the modified Newcastle-Ottawa Scale (NOS)[32, 33] for the representativeness of sample, appropriateness of sample size, response rate, validity of method, strategy to control confounding factors, reliability of outcome determination, and appropriate statistical analyses. The quality score (Table 2) disagreements were resolved by consensus and a final agreed-upon rating was assigned to each study (S2 file). Moreover, from the included studies each antibacterial tested for Klebsiella was extracted (Table 3).
Quality control
The quality of the included studies was checked independently by two authors (LG and KBT) using a set of predetermined criteria such as research design quality of paper, completeness of extractable information, and employed methods for Klebsiella species isolation.
Data analysis
A random-effects analysis method was employed to determine the pooled prevalence, subgroup analysis, and 95% confidence interval (CI) using the approach of Der- Simonian and Laird [34]. Variances and CIs were stabilized using Freeman-Tukey arc-sine methodology [35]. Heterogeneity of study results was assessed using I2 test and significant heterogeneity was considered at p < 0.10 and I2 > 50 [34, 36]. Open Meta-Analyst (version 3.13) and Comprehensive Meta-Analysis (version 3.1) statistical analyses were used. Moreover, subgroup analyses based on administrative regions and mechanism of antibacterial action were also performed to improve the specificity of the assessment of the tested drugs.
Result
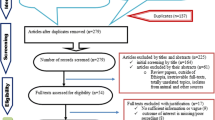
We obtained 174 potentially relevant studies through searching electronic databases. From these, 88 duplicate articles were removed by the help of Endnote(version X7)[Niles software, Clarivate analytic company); and the remaining 86 records were screened using their title and abstract out of which 35 articles were omitted based on the exclusion criteria. Full texts of 51 records were evaluated for eligibility in which 16 articles were removed due to data incompleteness, small number of isolates and/or tested antibiotics (less than 5 Klebsiella isolates and antibiotics in a single study). Finally 35 eligible studies were included for the meta-analysis (Fig. 1).
Cross-sectional study design was used in all the included 35 studies. A total of 13,269 study samples were employed, from which 1017 Klebsiella species were isolated. The included studies were taken from Amhara, Oromia. Southern Nations, Nationalities and People (SNNP), Tigray and Addis Ababa; however, there was no study obtained from other regions of Ethiopia (Afar, Benishangul-Gumuz, Gambella, Somali, Harari and Dire Dawa city administration). For screening of Klebsiella species, various specimens were utilized from the various part of the human body, including blood, urine, sputum, body fluids, ear discharge, wound swab, eye swab, stools and pus (Table 2).
The paper-based analysis of this study showed that the overall Klebsiella resistance in Ethiopia was 53.75% (95% CI: 48.35—58.94%) (Fig. 2). Subgroup analyses were carried out based on the region (Addis Ababa, Amhara, Oromia, SNNP, and Tigray), and then the average prevalence of Klebsiella resistance was determined in region wise. Southern Nations, Nationalities, and Peoples of Ethiopia was ranked first (64.39%, 95% CI: 54.94–73.83%), followed by Addis Ababa (55.67%, 95% CI: 47.74–63.40%), Oromia (55.24%, 95% CI: 43.95–66.50%), Amhara (50.1%, 95% CI: 40.82–59.20%), whereas relatively low (but highly varied) prevalence of Klebsiella resistance was reported from Tigray region (46.16%, 95% CI: 21.97–70.34%) (Figs. 3 and 4). The level of heterogeneity of the included studies was high, by random model methods (I2 = 90.55%; P < 0.01).
Additional subgroup analyses were done to estimate the pooled prevalence of Klebsiella resistance based on the mechanism of action of the antibacterial drugs. Antimetabolite (a single drug, trimethoprim-sulfamethoxazole) accounted the highest resistance percentage (66.91%, 95% CI: 59.79–74.06%), followed by cell wall synthesis inhibitors (61.61%, 95% CI: 44.79–79.42%), protein synthesis inhibitors (45.95%, 95% CI: 27.29–64.62%), whereas nucleic acid synthesis inhibitors showed relatively low resistance percentage (35.98%, 95% CI: 31.83–40.14%) and level of heterogeneity of this class was not significant (I2 = 0%, p = 0.68). With regarding to individual antibiotics, high resistance rates were observed to ampicillin (90.56%, 95% CI: 86.31–94.81%), followed by amoxicillin (76.01%, 95% CI: 61.44–90.38%), trimethoprim-sulfamethoxazole (66.91%, 95% CI: 59.76–74.06%), ceftazidime (65.07%, 95% CI: 52.68–77.46%). A relatively low level of resistance rate was observed to amikacin (16.74, 95% CI: 6.84–26.64), and cefoxitin 29.73%, 95% CI: 12.05–47.41%) (Fig. 5).
Discussion
The nationwide meta-analysis of Klebsiella resistance was not done in Ethiopia so far. So we conducted a comprehensive systematic review and meta-analysis to determine the pooled of Klebsiella antimicrobial resistance in Ethiopia. From our findings Klebsiella displayed diverse resistance patterns, with percentages varying slightly based on the antibiotic type and geographical distribution. Based on this meta-analysis, the overall Klebsiella resistance in Ethiopia was found to be 53.75% (95% CI: 48.35—58.94%). Similar findings were reported from meta-analysis study done in sub-Saharan Africa and Iran, where the resistance of Klebsiella for some antibiotics was greater than 50% [17, 20]. This indicates more than half of the isolated Klebsiella species were resistance to the most common antibacterial drugs. The possible reasons could be over/under dose of antimicrobial drugs in treating infectious diseases, and empiric use of antibiotics which would inevitably lead to emergence of resistance [24].
Antimicrobial resistance is becoming a public health challenge in the 21st century as a result of many factors including misuse of antimicrobials by health professional and patients, inadequate surveillance systems [7, 27]. Particularly, the emergence and spread of resistant strains of Klebsiella species are a considerable threat to public health [37]. For Klebsiella, resistance to antimicrobials is a normal evolutionary process, but the resistance rate is frequently aggravated by misuse of antibiotics [13, 38]. As per previous study, from the different species of Klebsiella, Klebsiella pneumoniae is becoming known for its resistance properties to most of the last-line antibiotics [8].
In this meta-analysis, the subgroup analysis of regional prevalence of Klebsiella indicated that the highest prevalence of Klebsiella resistance (64.39%) was estimated in SNNP, which is relatively higher than Tigray region (46.16%) (Fig. 4). The observed variation might be due to differences in study location, hospital setup, and antimicrobial utilization. The level of heterogeneity of the included studies was high, by random model methods (I2 = 90.55%; P < 0.01). This indicates that, the included studies have been done in different study areas, study periods, and study populations, which could have an effect on the heterogeneity of the studies.
The other subgroup analysis, based on the mechanism of action of antimicrobials, showed that Klebsiella exhibited higher resistance to antimetabolites (66.91%), which is actually represented by single drug (trimethoprim-sulfamethoxazole), followed by cell wall synthesis inhibitors (61.61%)(Fig. 5). In our finding, there was only one drug (trimethoprim-Sulfamethoxazole) from the antimetabolites tested for Klebsiella resistance, which might have led to the highest value of resistance group compared to the others. However, the highest resistance rate of Klebsiella was noted from individual antibiotics grouped under cell wall synthesis inhibitors including ampicillin (90.56%), amoxicillin (76.01%) and amoxicillin-clavulanic acid (56.92%).
Our finding was concordant with studies done in West Africa where Klebsiella was resistant to Ampicillin (92.5%) and amoxicillin-clavulanic acid (66.1%) [39], and a little bit lower Klebsiella resistance to ampicillin was reported from the three districts of Uganda (66.5%) [40] and India [1]. The higher resistance rate of Klebsiella species to ampicillin and amoxicillin could possibly be due to the fact that the bacteria naturally produces extended spectrum beta-lactamases which inactivate beta lactam` antibiotics (ESBLs) [11, 20]. Our finding showed almost similar pooled Klebsiella resistance to ceftriaxone (47.6%) with previous meta-analysis done on wound infection in which more than half of Klebsiella pneumoniae isolates exhibited resistance to ceftriaxone (57%) [23].
The findings of this study indicated that there is high magnitude of Klebsiella antimicrobial resistance in Ethiopia. The possible reasons could be limited infection surveillance programs, the lack of communication between physicians and microbiologists, lack of standardized criteria to determine drug resistant isolates, limited laboratory facilities, and poor sanitation [20]. The relatively high rates of drug resistant isolates of Klebsiella seen in this meta-analysis might have negative effects on public health which could cause difficulty in treating Klebsiella pneumonia associated infections since only fewer effective drugs are available for treating these highly drug-resistant strains [20]. Hence, functional infection control committee, applying infection prevention protocols, advocating rational prescribing habits, appropriate antimicrobial therapy, health education; and improvement of personal and environmental hygiene need to be applied to curb the resistance problem [13, 18, 20, 41].
Some limitations in our study should be acknowledged. The pooled resistance of Klebsiella was not calculated based on species basis as there was shortage of studies done on individual species. Many of the included studies describe Klebsiella pneumonia, but rarely for other species. Therefore, this meta-analysis should be seen in the context of such limitations.
Conclusion
In this systematic review and meta-analysis, the pooled Klebsiella resistance was found to be considerably high (53.75%) to most of the essential antibiotics in Ethiopia. Klebsiella was highly resistant to ampicillin and amoxicillin but relatively lower to amikacin. Therefore, implementing proper antibiotic prescription policies and appropriate antimicrobial therapy could be the potential interventional strategies to address the emerging resistance of Klebsiella species.
Data Availability
There is no remaining data and materials; all information is clearly presented in the main manuscript.
Abbreviations
- AML:
-
Amoxicillin
- AMC:
-
Amoxicillin-clavulanic acid
- AMP:
-
Ampicillin
- C:
-
Chloramphenicol
- CIP:
-
Ciprofloxacin
- GEN:
-
Gentamicin
- CRO:
-
Ceftriaxone
- F:
-
Nitrofurantoin
- NA:
-
Nalidixic acid
- NOR:
-
Norfoxacin
- SXT:
-
Trimethoprim/Sulfamethoxazole
- TE:
-
Tetracycline
References
Perween N, Prakash SK, Siddiqui O. Multi drug resistant Klebsiella isolates in burn patients: a comparative study. J Clin Diagn Research: JCDR. 2015;9(9):DC14.
Abayneh M, Tesfaw G, Abdissa A. Isolation of extended-spectrum β-lactamase-(ESBL-) producing Escherichia coli and Klebsiella pneumoniae from patients with community-onset urinary tract infections in Jimma University Specialized Hospital, Southwest Ethiopia. Canadian Journal of Infectious Diseases and Medical Microbiology. 2018;2018.
Ameshe A, Engda T, Gizachew M. Prevalence, antibiotic susceptibility patterns, including ESBL production and associated risk factors of Klebsiella species among UTI suspected patients at Bahir-dar town, Northwest Ethiopia. 2020.
Xu L, Sun X, Ma X. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann Clin Microbiol Antimicrob. 2017;16(1):1–12.
Lam MM, Wyres KL, Duchêne S, Wick RR, Judd LM, Gan Y-H, et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun. 2018;9(1):1–10.
Mohammed A, Seid ME, Gebrecherkos T, Tiruneh M, Moges F. Bacterial isolates and their antimicrobial susceptibility patterns of wound infections among inpatients and outpatients attending the University of Gondar Referral Hospital, Northwest Ethiopia. International Journal of Microbiology. 2017;2017.
Sakkas H, Bozidis P, Ilia A, Mpekoulis G, Papadopoulou C. Antimicrobial Resistance in bacterial pathogens and detection of Carbapenemases in Klebsiella pneumoniae isolates from Hospital Wastewater. Antibiotics. 2019;8(3):85.
Effah CY, Sun T, Liu S, Wu Y. Klebsiella pneumoniae: an increasing threat to public health. Ann Clin Microbiol Antimicrob. 2020;19(1):1–9.
Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41(3):252–75.
Heidary M, Goudarzi H, Hashemi A, Eslami G, Goudarzi M, Chirani A et al. The prevalence of genes that encode quinolone resistance in Klebsiella pneumoniae strains isolated from hospitalized patients during 2013–2014. Arch Pediatr Infect Dis. 2016.
El Bouamri M, Arsalane L, El Kamouni Y, Zouhair S. Antimicrobial susceptibility of urinary Klebsiella pneumoniae and the emergence of carbapenem-resistant strains: a retrospective study from a university hospital in Morocco, North Africa. Afr J Urol. 2015;21(1):36–40.
Perovic O, Singh-Moodley A, Duse A, Bamford C, Elliott G, Swe-Han KS, et al. National sentinel site surveillance for antimicrobial resistance in Klebsiella pneumoniae isolates in South Africa, 2010–2012. South Afr Med J. 2014;104(8):563–8.
Tuem KB, Gebre AK, Atey TM, Bitew H, Yimer EM, Berhe DF. Drug resistance patterns of Escherichia coli in Ethiopia: a meta-analysis. BioMed research international. 2018;2018.
Laxminarayan R, Matsoso P, Pant S, Brower C, Røttingen J-A, Klugman K, et al. Access to effective antimicrobials: a worldwide challenge. The Lancet. 2016;387(10014):168–75.
Asante J, Osei Sekyere J. Understanding antimicrobial discovery and resistance from a metagenomic and metatranscriptomic perspective: advances and applications. Environ Microbiol Rep. 2019;11(2):62–86.
Mbelle NM, Osei Sekyere J, Amoako DG, Maningi NE, Modipane L, Essack SY, et al. Genomic analysis of a multidrug-resistant clinical Providencia rettgeri (PR002) strain with the novel integron ln1483 and an A/C plasmid replicon. Ann N Y Acad Sci. 2020;1462(1):92–103.
Lester R, Musicha P, Van Ginneken N, Dramowski A, Hamer DH, Garner P, et al. Prevalence and outcome of bloodstream infections due to third-generation cephalosporin-resistant Enterobacteriaceae in sub-saharan Africa: a systematic review. J Antimicrob Chemother. 2020;75(3):492–507.
Muhie OA. Antibiotic use and resistance pattern in ethiopia: systematic review and meta-analysis. International journal of microbiology. 2019;2019.
Ferreira RL, da Silva B, Rezende GS, Nakamura-Silva R, Pitondo-Silva A, Campanini EB, et al. High prevalence of multidrug-resistant Klebsiella pneumoniae harboring several virulence and β-lactamase encoding genes in a brazilian intensive care unit. Front Microbiol. 2019;9:3198.
Heidary M, Nasiri MJ, Dabiri H, Tarashi S. Prevalence of drug-resistant Klebsiella pneumoniae in Iran: a review article. Iran J Public Health. 2018;47(3):317.
Shatalov A. Prevalence and antibiotic resistance pattern of Escherichia coli and Klebsiella pneumoniae in urine tract infections at the La Paz Medical center, Malabo, Equatorial Guinea. Open J Med Microbiol. 2015;5(04):177.
Leopold SJ, van Leth F, Tarekegn H, Schultsz C. Antimicrobial drug resistance among clinically relevant bacterial isolates in sub-saharan Africa: a systematic review. J Antimicrob Chemother. 2014;69(9):2337–53.
Sisay M, Worku T, Edessa D. Microbial epidemiology and antimicrobial resistance patterns of wound infection in Ethiopia: a meta-analysis of laboratory-based cross-sectional studies. BMC Pharmacol Toxicol. 2019;20(1):35.
Dagnew M, Yismaw G, Gizachew M, Gadisa A, Abebe T, Tadesse T, et al. Bacterial profile and antimicrobial susceptibility pattern in septicemia suspected patients attending Gondar University Hospital, Northwest Ethiopia. BMC Res Notes. 2013;6(1):283.
Diriba K, Kassa T, Alemu Y, Bekele S. In Vitro Biofilm Formation and Antibiotic Susceptibility Patterns of Bacteria from Suspected External Eye Infected Patients Attending Ophthalmology Clinic, Southwest Ethiopia. International Journal of Microbiology. 2020;2020.
Merga Duffa Y, Terfa Kitila K, Mamuye Gebretsadik D, Bitew A. Prevalence and antimicrobial susceptibility of bacterial uropathogens isolated from pediatric patients at yekatit 12 hospital medical college, Addis Ababa, Ethiopia. International journal of microbiology. 2018;2018.
Eshetie S, Unakal C, Gelaw A, Ayelign B, Endris M, Moges F. Multidrug resistant and carbapenemase producing Enterobacteriaceae among patients with urinary tract infection at referral hospital, Northwest Ethiopia. Antimicrob Resist Infect Control. 2015;4(1):12.
Eshetie S, Tarekegn F, Moges F, Amsalu A, Birhan W, Huruy K. Methicillin resistant Staphylococcus aureus in Ethiopia: a meta-analysis. BMC Infect Dis. 2016;16(1):689.
Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Reprint—preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Phys Ther. 2009;89(9):873–80.
Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JP, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. J Clin Epidemiol. 2009;62(10):e1–e34.
Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Int J Surg. 2021;88:105906.
Wells GA, Shea B, O’Connell D, Peterson J, Welch V, Losos M et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Oxford; 2000.
Peterson J, Welch V, Losos M, Tugwell P. The Newcastle-Ottawa scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Ottawa: Ottawa Hospital Research Institute; 2011.
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–88.
Fazel S, Khosla V, Doll H, Geddes J. The prevalence of mental disorders among the homeless in western countries: systematic review and meta-regression analysis. PLoS Med. 2008;5(12):e225.
Rücker G, Schwarzer G, Carpenter JR, Schumacher M. Undue reliance on I 2 in assessing heterogeneity may mislead. BMC Med Res Methodol. 2008;8(1):79.
Harada K, Shimizu T, Mukai Y, Kuwajima K, Sato T, Usui M, et al. Phenotypic and molecular characterization of antimicrobial resistance in Klebsiella spp. isolates from companion animals in Japan: clonal dissemination of multidrug-resistant extended-spectrum β-lactamase-producing Klebsiella pneumoniae. Front Microbiol. 2016;7:1021.
Laxminarayan R, Duse A, Wattal C, Zaidi AK, Wertheim HF, Sumpradit N, et al. Antibiotic resistance—the need for global solutions. Lancet Infect Dis. 2013;13(12):1057–98.
Bernabe KJ, Langendorf C, Ford N, Ronat J-B, Murphy RA. Antimicrobial resistance in West Africa: a systematic review and meta-analysis. Int J Antimicrob Agents. 2017;50(5):629–39.
Najjuka CF, Kateete DP, Kajumbula HM, Joloba ML, Essack SY. Antimicrobial susceptibility profiles of Escherichia coli and Klebsiella pneumoniae isolated from outpatients in urban and rural districts of Uganda. BMC Res Notes. 2016;9(1):235.
Zhang Y, Ma Y, Ye L, Luo Y, Yang J. Prevalence and antimicrobial susceptibility of hypervirulent Klebsiella pneumoniae isolates in China. Clin Infect Dis. 2014;58(10):1493–4.
Abebaw A, Tesera H, Belachew T, Mihiretie GD. The bacterial profile and antibiotic susceptibility pattern among patients with suspected bloodstream infections, Gondar, north-west Ethiopia. Pathol Lab Med Int. 2018;10:1–7.
Abebe M, Tadesse S, Meseret G, Derbie A. Type of bacterial isolates and antimicrobial resistance profile from different clinical samples at a Referral Hospital, Northwest Ethiopia: five years data analysis. BMC Res Notes. 2019;12(1):568.
Adhanom G, Gebreegziabiher D, Weldu Y, Gebreyesus Wasihun A, Araya T, Legese H et al. Species, risk factors, and antimicrobial susceptibility profiles of bacterial isolates from HIV-infected patients suspected to have pneumonia in Mekelle zone, Tigray, northern Ethiopia. BioMed research international. 2019;2019.
Alemayehu T, Ali M, Mitiku E, Hailemariam M. The burden of antimicrobial resistance at tertiary care hospital, southern Ethiopia: a three years’ retrospective study. BMC Infect Dis. 2019;19(1):585.
Amsalu A, Geto Z, Asegu D, Eshetie S. Antimicrobial resistance pattern of bacterial isolates from different clinical specimens in Southern Ethiopia: a three year retrospective study. Afr J Bacteriol Res. 2017;9(1):1–8.
Awoke N, Kassa T, Teshager L. Magnitude of biofilm formation and antimicrobial resistance pattern of bacteria isolated from urinary catheterized inpatients of Jimma University medical center, Southwest Ethiopia. International journal of microbiology. 2019;2019.
Beyene D, Bitew A, Fantew S, Mihret A, Evans M. Multidrug-resistant profile and prevalence of extended spectrum β-lactamase and carbapenemase production in fermentative gram-negative bacilli recovered from patients and specimens referred to National Reference Laboratory, Addis Ababa, Ethiopia. PLoS ONE. 2019;14(9):e0222911.
Bitew A, Molalign T, Chanie M. Species distribution and antibiotic susceptibility profile of bacterial uropathogens among patients complaining urinary tract infections. BMC Infect Dis. 2017;17(1):1–8.
Dereje M, Woldeamanuel Y, Asrat D, Ayenachew F. Urinary tract infection among fistula patients admitted at Hamlin fistula hospital, Addis Ababa, Ethiopia. BMC Infect Dis. 2017;17(1):150.
Dessalegn L, Shimelis T, Tadesse E, Gebre-selassie S. Aerobic bacterial isolates from post-surgical wound and their antimicrobial susceptibility pattern: a hospital based cross-sectional study. J Med Res. 2014;3(2):18–23.
Feleke T, Eshetie S, Dagnew M, Endris M, Abebe W, Tiruneh M, et al. Multidrug-resistant bacterial isolates from patients suspected of nosocomial infections at the University of Gondar Comprehensive Specialized Hospital, Northwest Ethiopia. BMC Res Notes. 2018;11(1):602.
Gashaw M, Berhane M, Bekele S, Kibru G, Teshager L, Yilma Y, et al. Emergence of high drug resistant bacterial isolates from patients with health care associated infections at Jimma University medical center: a cross sectional study. Antimicrob Resist Infect Control. 2018;7(1):138.
Gebremariam G, Legese H, Woldu Y, Araya T, Hagos K, GebreyesusWasihun A. Bacteriological profile, risk factors and antimicrobial susceptibility patterns of symptomatic urinary tract infection among students of Mekelle University, northern Ethiopia. BMC Infect Dis. 2019;19(1):950.
Getahun E, Gelaw B, Assefa A, Assefa Y, Amsalu A. Bacterial pathogens associated with external ocular infections alongside eminent proportion of multidrug resistant isolates at the University of Gondar Hospital, northwest Ethiopia. BMC Ophthalmol. 2017;17(1):151.
Godebo G, Kibru G, Tassew H. Multidrug-resistant bacterial isolates in infected wounds at Jimma University Specialized Hospital, Ethiopia. Ann Clin Microbiol Antimicrob. 2013;12(1):17.
Gutema T, Weldegebreal F, Marami D, Teklemariam Z. Prevalence, antimicrobial susceptibility pattern, and associated factors of urinary tract infections among adult diabetic patients at Metu Karl Heinz Referral Hospital, Southwest Ethiopia. International Journal of Microbiology. 2018;2018.
Hailu D, Derbie A, Mekonnen D, Zenebe Y, Adem Y, Worku S, et al. Drug resistance patterns of bacterial isolates from infected wounds at Bahir Dar regional Health Research Laboratory center, Northwest Ethiopia. Ethiop J Health Dev. 2016;30(3):112–7.
Legese MH, Weldearegay GM, Asrat D. Extended-spectrum beta-lactamase-and carbapenemase-producing Enterobacteriaceae among ethiopian children. Infect drug Resist. 2017;10:27.
Mama M, Abdissa A, Sewunet T. Antimicrobial susceptibility pattern of bacterial isolates from wound infection and their sensitivity to alternative topical agents at Jimma University Specialized Hospital, South-West Ethiopia. Ann Clin Microbiol Antimicrob. 2014;13(1):14.
Mamuye Y. Antibiotic resistance patterns of common Gram-negative uropathogens in St. Paul’s Hospital Millennium Medical College. Ethiop J Health Sci. 2016;26(2):93–100.
Mengesha RE, Kasa BG-S, Saravanan M, Berhe DF, Wasihun AG. Aerobic bacteria in post surgical wound infections and pattern of their antimicrobial susceptibility in Ayder Teaching and Referral Hospital, Mekelle, Ethiopia. BMC Res Notes. 2014;7(1):1–6.
Mitiku E, Amsalu A, Tadesse BT. Pediatric urinary tract infection as a cause of outpatient clinic visits in southern Ethiopia: a cross sectional study. Ethiop J Health Sci. 2018;28(2):187–96.
Feleke Moges SE, Wondwossen Abebe F, Mekonnen, Mulat Dagnew AE, Azanaw Amare T, Feleke M, Gizachew TM. High prevalence of extended-spectrum betalactamase-producing Gram-negative pathogens from patients attending Felege Hiwot Comprehensive Specialized Hospital, Bahir Dar, Amhara region. PLoS ONE. 2019;14(6):1–13.
Molla R, Tiruneh M, Abebe W, Moges F. Bacterial profile and antimicrobial susceptibility patterns in chronic suppurative otitis media at the University of Gondar Comprehensive Specialized Hospital, Northwest Ethiopia. BMC Res Notes. 2019;12(1):414.
Negussie A, Mulugeta G, Bedru A, Ali I, Shimeles D, Lema T, et al. Bacteriological profile and antimicrobial susceptibility pattern of blood culture isolates among septicemia suspected children in selected hospitals Addis Ababa, Ethiopia. Int J Biol Med Res. 2015;6(1):4709.
Sahile T, Esseye S, Beyene G, Ali S. Post-surgical infection and antibiotic susceptibility patterns of bacteria isolated from admitted patients with signs of infection at Jimma University specialized hospital, Jimma, Ethiopia. Int J Trop Dis Health. 2016;17(4):1–12.
Sorsa A, Früh J, Stötter L, Abdissa S. Blood culture result profile and antimicrobial resistance pattern: a report from neonatal intensive care unit (NICU), Asella teaching and referral hospital, Asella, south East Ethiopia. Antimicrob Resist Infect Control. 2019;8(1):42.
Tadesse B, Shimelis T, Worku M. Bacterial profile and antibacterial susceptibility of otitis media among pediatric patients in Hawassa, Southern Ethiopia: cross-sectional study. BMC Pediatr. 2019;19(1):398.
Tadesse S, Kahsay T, Adhanom G, Kahsu G, Legese H, Derbie A. Prevalence, antimicrobial susceptibility profile and predictors of asymptomatic bacteriuria among pregnant women in Adigrat General Hospital, Northern Ethiopia. BMC Res Notes. 2018;11(1):740.
Teklu DS, Negeri AA, Legese MH, Bedada TL, Woldemariam HK, Tullu KD. Extended-spectrum beta-lactamase production and multi-drug resistance among Enterobacteriaceae isolated in Addis Ababa, Ethiopia. Antimicrob Resist Infect Control. 2019;8(1):39.
Wasihun AG, Zemene Y. Bacterial profile and antimicrobial susceptibility patterns of otitis media in Ayder Teaching and Referral Hospital, Mekelle University, Northern Ethiopia. SpringerPlus. 2015;4(1):701.
Worku S, Gelaw A, Aberra Y, Muluye D, Derbie A, Biadglegne F. Bacterial etiologies, antibiotic susceptibility patterns and risk factors among patients with ear discharge at the University of Gondar Hospital, Northwest Ethiopia. Asian Pac J Trop Dis. 2017;7(1):36–42.
Acknowledgements
We don’t have any person or organization to acknowledge.
Funding
There was no any fund.
Author information
Authors and Affiliations
Contributions
LG contributed to conception of the research and study design. LG, TT, GGK and KBT have contributed to literature review, data collection data extraction. All authors made substantial contributions to analysis and interpretation of data; and took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Disclosure
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Gebremeskel, L., Teklu, T., Kasahun, G.G. et al. Antimicrobial resistance pattern of Klebsiella isolated from various clinical samples in Ethiopia: a systematic review and meta-analysis. BMC Infect Dis 23, 643 (2023). https://doi.org/10.1186/s12879-023-08633-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-023-08633-x