Abstract
Background
During the last six decades, extensive use of antibiotics has selected resistant strains, increasing the rate of fatal infectious diseases, and exerting an economic burden on society. This situation is widely accepted as a global problem, yet its degree is not well elucidated in many regions of the world. Up till now, no systemic analysis of Antimicrobial resistance (AMR) in Pakistan has been published. The current study aims to describe the antibiotic-resistance scenario of Pakistan from human samples of the last 10 y, to find the gaps in surveillances and methodology and recommendations for researchers and prescribers founded on these outcomes.
Methods
Original research articles analyzed the pattern of Antibiotic resistance of any World Health Organization (WHO) enlisted priority pathogens in Pakistan (published onward 2009 till March 2020), were collected from PubMed, Google scholar, and PakMedi Net search engines. These articles were selected based on predefined inclusion and exclusion criteria. Data about the study characteristics and antibiotic-resistance for a given bacterium were excluded from literature. Antibiotic resistance to a particular bacterium was calculated as a median resistance with 95% Confidence Interval (CI).
Results
Studies published in the last 10 y showed that Urinary Tract Infection (UTI) is the most reported clinical diagnosis (16.1%) in Pakistan. E. coli were reported in 28 (30.11%) studies showing high resistance to antibiotics’ first line. Methicillin-resistant Staphylococcus aureus (MRSA) was found in 49% of S. aureus’ total reported cases. Phenotypic resistance pattern has mostly been evaluated by Disk Diffusion Method (DDM) (82.8%), taken Clinical Laboratory Standards Institute (CLSI) as a breakpoint reference guideline (in 79.6% studies). Only 28 (30.11%) studies have made molecular identification of the resistance gene. blaTEM (78.94% in Shigella spp) and blaNDM-1 (32.75% in Klebsiella spp) are the prominent reported resistant genes followed by VanA (45.53% in Enterococcus spp), mcr-1 (1.61% in Acinetobacter spp), and blaKPC-2 (31.67% in E. coli). Most of the studies were from Sindh (40.86%), followed by Punjab (35.48%), while Baluchistan’s AMR data was not available.
Conclusion
Outcomes of our study emphasize that most of the pathogens show high resistance to commonly used antibiotics; also, we find gaps in surveillances and breaches in methodological data. Based on these findings, we recommend the regularization of surveillance practice and precise actions to combat the region’s AMR.
Similar content being viewed by others
Background
Antibiotic-resistance is the ability of bacteria to be not cured or prevented by the antibiotics used against them. Ever since, from the start of antibiotic development, there was a continuous worry about the resistance of bacteria to antibiotics. It is one of the significant hazards developed by bacteria because it not only causes deadly infections but also bases extended illness, high budget outlay, and increased morbidity. The poor management, unhygienic environment, untrained professionals, overuse, and misuse of antibiotics are the factors that lead to the development of theses panic situations in the form of adopting or acquiring resistant genes by bacteria [1]. The World Health Organization personifies antimicrobial resistance as a public health emergency that must be coped with the supreme insistence [2].
AMR is a serious issue worldwide, especially in less developed countries. South-Asia is deliberated to be the central region for antibiotic-resistant bacteria. It is anticipated that 70% of antibiotic resistance is ascending in the Asia region, making it county-wide and worldwide hazard [3]. Pakistan is a developing country of the South-Asia, rich in antibiotic resistance, a significant global and regional threat [4]. Both the multi-drug resistant (MDR) and extensively drug resistant (XDR) bacteria are identified in Pakistan in the last few years. In the last decade from Pakistan, resistance against quinolones has increased for Enterobacteriaceae [5]. In 2016, the outbreak of XDR Salmonella was one of its examples that show even 100% resistance to fluoroquinolones [6].
Similarly, a blood stream infection (BSI) study shows even 93.7% resistant isolates to third-generation cephalosporin [7]. The high prevalence of Metallo-β-lactamase (MBL) up to 71% and Extended Spectrum β-Lactamase (ESBL) up to 40%, carbapenem-resistant bacteria-harboring blaNDM, blaKPC genes, and the mcr-1 gene that show resistance to colistin, the last drug of choice, are reported from human isolates [4, 8,9,10]. Regarding these findings, we are on the edge of antibiotic therapy. The reason behind this is demonstrated in various studies, which are irrational prescribing, incentives for overprescribing, self-medication, unqualified staff, lack of formal training, nonentity of culture sensitivity tests, and the incomplete dosage taken by patients [11].
Numerous individual studies are accomplished on the prevalence of AMR in Pakistan. However, no such a systematical report is published to present a comprehensive depiction of antibiotic resistance in Pakistan. In this study, we aim to amalgamate the rate of antibiotic resistance in clinically substantial bacteria from Pakistan. Our alternative goal is to find out the slits in surveillance, reference for imminent work, to offer sanctions and guides for officials and prescribers for indication founded approaches towards mitigating AMR in Pakistan.
Methods
Literature search
The guidelines of Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) were followed to accomplish this systematic review. Research articles were searched out on PubMed, Google scholar, and PakMedi Net search engines by giving them pertinent keywords like antimicrobial resistance, antibiotic, resistance, resistant, susceptible, pathogens (also specifying pathogen name) in Pakistan, published onward 2009 till dated March 2020. Initially, the literature was selected from the title and abstract. The duplicate was removed and further filtered out by reviewing the whole text considering inclusion and exclusion criteria (Fig. 1).
Inclusion criteria:
-
Studies having at least 30 bacteria, isolated from human samples in Pakistan (according to the Central Limit Theorem, the minimum sample size is 30),
-
Articles published onward 2009 till March 2020 in the English language,
-
Studied AMR of any WHO enlisted priority bacterial pathogen [12] from Pakistan.
-
Studies done in laboratory site with confident cutoff value for antimicrobial sensitivity testing.
-
Mentioned the total sample size and the resistance/susceptible percentage of bacteria.
Exclusion criteria
Studies having bacterial isolates from environmental, animal, or poultry origin, Published before 2009, bacterial isolates less than 30, not mentioned the antibiotic-resistant profile, reviews, language other than English, and articles that not used the standard methods.
Data mining
The selected studies were evaluated to gather the data on the duration of the study, year of publication, location of the study, patients type, samples type, clinical diagnosis, gender, age group, samples size, bacterial identification methods, bacterial type, quantitative antibiotics resistance pattern, antibiotic resistance detection methods, breakpoint reference guidelines, and antibiotic resistance genes, from each study on Excel Sheet 2016. Data extractions were performed by two researchers HB and MNK, separately to negotiate any possible errors.
Data analysis
The articles for systematic review analysis were selected considering the inclusion and exclusion criteria. All the data about the study characteristics were determined considering the authenticity of evaluation methods. Patients were having an age of less than one month considered as neonates, less than 18 years as pediatric, and above 18 years as adults. The intermediate resistances were considering as resistance in this study. Each bacterium’s antibiotic resistance profile for every antibiotic was determined in the form of Median resistance (MR) with 95% confidence interval (CI) to compute a standardized measure for collective data. Statistical analysis and visualization of data were performed using Microsoft excel 2016, GraphPad Prism 8.0.2, and Inkscape 0.92.4.
Results
Literatures features
A total of 93 articles were selected for systematic analysis considering the inclusion and exclusion criteria out of 216 articles collected from search engines based on the keywords. Out of 93 articles 64 were from gram negative bacteria [6, 9, 10, 13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73], 16 were from gram positive bacteria [74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89], and 13 had data about both gram positive and gram negative bacteria [90,91,92,93,94,95,96,97,98,99,100,101,102].
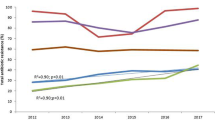
Pakistan consists of four provinces and capital territory, i-e, Islamabad. Most studies (31.6%) were reported from Karachi (Sindh), followed by Lahore (Punjab) 16.7%. 11.95% of studies were reported from Khyber Pakhtunkhwa province and 10.86% from the Islamabad region. However, no study was reported from Baluchistan province, only in one study reported from Karachi, 4% of total samples were from Baluchistan [89]. In one study, the province or city was not mentioned, while in one study from Punjab province, the city name was not mentioned (Fig. 2) [31, 96]. The maximum number of studies were reported in 2016 (13.9%), followed by 2019 (13%). 39.1% of studies have sample collection duration in the range of 2009 to 2014, while in 7 studies; the date and duration of sample collection were not mentioned. The numbers of studies based on the year of publication and sampling duration are stated in (Fig. 3).
Phenotypic detection of antibiotic resistance by DDM was reported in 82.8% of the total studies and 79.6% of total studies used CLSI as the breakpoint reference guidelines. UTI was the most testified clinical diagnosis, 16.1% of the total studies, while 36.6% of total studies were not declared about the clinical diagnosis. Among the data from Urinary tract infections, two studies stated the community acquired UTI [27, 93] and one study demonstrates both hospital and community acquired UTI [30]. However the remaining articles did not mention about the source of UTI [25, 26, 34, 35, 39, 41, 42, 49, 53, 76, 94]. Data about the clinical diagnosis concerning bacterial pathogens are mentioned in Table 1. The E. coli were documented in 28 studies; however, we did not find any study on Enterobacter cloacae, Campylobacter jejuni, and Serratia marcescens according to our inclusion criteria. 32.5% of total studies include inpatient samples, while 28.8% of studies were not specified about the patient type. The mean ages were reported in 18 (19.35%) studies i-e (Median 48.32, 95%CI: 29.58–53.98). 41 (44.08%) and 31 (33.33%) of studies had no information about the age group and gender. Data about the source of infection were not available in 82.79% of the total studies (Table 2). The number and percentage of studies regarding study characteristics i-e patient type, gender, age groups bacterial identification method, phenotypic detection method, and break point reference guidelines for gram positive and gram negative bacteria are presented in Table 3, Table 4, Table 5, and Table 6. Data about the studies used gram staining and different conventional biochemical testing for bacterial identification are presented in Table 7.
Antibiotic-resistance/susceptible pattern
The MR with 95%CI was calculated for ten bacteria. However, due to insufficient available data of N. gonorrhoeae and H. influenzae, their MR was not considered. The MDR bacteria were reported in 8 (8.60%) studies, while 2 (2.15%) studies reported XDR bacteria (Table 8).
The E. coli were reported in 28 studies, showing high resistance to penicillin (MR 100, 95% CI: 82–100), cephradine (MR 92, 95%CI: 74–95), ampicillin (MR 90.55, 95%CI: 83–96%), and amoxicillin (MR 85, 95%CI: 33–100). Nevertheless, they are 100% susceptible to colistin, 94.5% to cefoperazone-sulbactam, 93.5% to imipenem, and 92% to meropenem. The complete antibiotic-resistant profile for E. coli is presented in (Fig. 4). Klebsiella spp. were reported in 13 studies showing high resistance to second and 3rd generation antibiotics i-e cefaclor (MR 100%) and cefotaxime (MR 82.5, 95% CI 22–100). However, they are susceptible to colistin (nearly 100%), imipenem (92%), and cefoperazone-sulbactam (91.5%) (Fig. 5). Proteus spp were reported in 2 studies showing high resistance to cefotaxime, ceftriaxone, and tobramycin which are (MR 66.5, 95% CI: 59–74), (MR 62.5, 95% CI: 49–76), and (MR 59.5, 95%CI: 36–83), respectively (Fig. 6). Salmonella spp. were reported in 10 studies from the Sindh region (Hyderabad, Karachi) during the last decade showing highly resistant to ciprofloxacin (MR 90.5, 95%CI: 12–100). However, they are 99–100% susceptible to ceftriaxone, imipenem, and meropenem (Fig. 7). Shigella spp. were reported in 4 studies showing the highest resistance to co-trimoxazole, and ampicillin i-e (MR 80, 95%CI: 56–85) and (MR 68, 95%CI: 4–68). According to reported studies, ofloxacin (MR 2.5%) and nalidixic acid (MR 3%) are among the most efficient antibiotics against Shigella spp. (Fig. 8).
H. pylori were reported in three studies showing high resistance to metronidazole (MR 89, 95%CI: 74–98) while 96 and 76% of species were susceptible to tetracycline and ofloxacin, respectively (Fig. 9).
Acinetobacter spp. were reported in 15 studies showing high resistance to almost all tested antibiotics except colistin, tigecycline, and minocycline, whose susceptibility was nearly 99.5%, 97.15, and 67% (Fig. 10). Pseudomonas spp. were reported in 13 studies showing high resistance to ceftazidime and aztreonam i-e (MR 73.5, 95%CI: 42–100) and (MR 70, 95%CI: 21–78). The resistance pattern for carbapenems i-e for meropenem were (MR 18 95%CI: 5–100) and imipenem were (MR 26.5 95%CI: 6–82). For piperacillin- tazobactam the MR were 18.5% against 1066 tested isolates. Moreover, it also shows high resistance to colistin i-e (MR 20, 95%CI: 0–41%). The highest susceptible among the tested antibiotics were for cefoperazone-sulbactam (86.5%). The complete depict of resistance profile of Pseudomonas spp. from the available data are presented in (Fig. 11).
S. aureus were reported in 20 studies in which 13 studies also report MRSA. S. aureus shows high resistance to penicillin followed by cefoxitin and levofloxacin i-e (MR 98, 95%C1: 95–100), (MR 83, 95%CI: 48–100%) and (MR 80, 95%CI: 56–85) respectively. Effective antibiotics against S. aureus were tigecycline, tetracycline, linezolid, and vancomycin, whose susceptibility was 100, 100, 99, and 98%, respectively, while 2% of S. aureus were VRSA (Fig. 12). Three studies determined MIC of vancomycin for S. aureus by different methods i-e by broth micro dilution and automated VITEK 2 system showing 100% susceptibility [79, 89]. However one study performed E test strip method showing 13 and 4.16% resistance to vancomycin for MRSA and MSSA respectively [80]. Enterococcus spp. were reported in 4 studies showing high resistant to oxacillin (MR 100%), and erythromycin (MR 96, 95%CI: 79–100), while linezolid showed 100% susceptibility against 240 tested isolates (Fig. 13).
Antibiotics resistance genes
Antibiotic-resistant genes were found out in 28 (30.11%) studies, in which two studies (7.14%) performed whole-genome sequencing [9, 22]. One study (3.57%) detects resistant genes via TaqMan® real-time PCR [37]. In comparison, twenty-five studies (89.28%) performed a conventional PCR assay for the investigation of resistant genes. None of the molecular studies were found in included literature of Neisseria gonorrhoeae, Haemophilus influenzae, Proteus spp., and Streptococcus pneumoniae. The complete depiction of resistant genes types, bacterial isolates, and investigated studies is presented in Table 9.
Discussion
Antibiotic-resistant is a global issue worldwide, but developing countries are more in threat because of less hygienic conditions and poor clinical infrastructure. The present study is the first systematic review from Pakistan to analyze the antibiotic-resistant data from the last ten years. In the present study, UTI was among the highest reported clinical diagnosis. In bacterial pathogen, E. coli was reported in the maximum number of studies showing high resistance to the first-line antibiotics. Similar is Bangladesh’s and Africa’s scenario due to the same trend of inappropriate antibiotics use in developing countries. However, resistance to levofloxacin and tetracycline is higher in the current study, which might be due to differences in AMR testing methodologies [103, 104]. Our data support the increasing trend of fluoroquinolone-resistant Salmonella spp. in the Asia region [105] as here we find (MR 90.5, 95%CI: 12–100) for ciprofloxacin out of 7392 tested isolates. However, the clinicians may prescribe cefixime, ceftriaxone, and carbapenem due to their significant reported susceptibility. In this study, Shigella spp. were reported in 4 studies showing MR 80% to co-trimoxazole. WHO classified Shigella spp. as the primary bacteria causing community-acquired infection [106]; therefore, more researches are required to get a deep insight. H. pylori show the highest resistance to metronidazole (MR 89, 95%CI 74–98%), which is more than Malaysia (82%) and China (77%). High resistance to metronidazole is due to its increased prescribing and easy availability in Pakistan [107]. We suggest more research work on the prevalence of antibiotic resistance targeting Neisseria gonorrhoeae, Haemophilus influenzae, Streptococcus pneumoniae, Serratia spp., Campylobacter spp., and Proteus spp. due to their less available data from Pakistan.
Acinetobacter spp. and Pseudomonas spp., which are intrinsically resistant to many antibiotics, also show a high rate of resistance to other CLSI recommended antibiotics like Acinetobacter spp. show MR 91.5% to meropenem and Pseudomonas spp. show MR 20% to colistin. The emerging trend is due to acquired resistance [108]. Our findings support 2017 WHO report in which they categorized Acinetobacter spp. and Pseudomonas spp. as critically priority bacteria [12].
MRSA is considering for high mortality rates [109]. In the current study, among 7469 tested S. aureus, 49% were MRSA. The actual value might be different due to the difference in the source of infection [110]. Vancomycin-resistant Enterococcus (VRE) is mainly involved in hospital-acquired infections [111]. In the current research, VRE was (MR 10.5, 95%CI: 1.8–100), which is more than Finland, Holland, Italy, Canada, and Bangladesh [103, 112]. The high incidence might be due to the VRE outbreak from an unknown source and the existence of vanA gene-encoded VRE reservoir in Pakistan [113].
The molecular antibiotic-resistant study is essential to get in-depth knowledge about the resistance mechanism (intrinsic or acquiring), which may help prevent and design novel or alternative therapeutic agents [114]. In the current study, 28(30.11%) studies reported the antibiotic-resistant genes in which the most prominent are ESBL and carbapenem-resistant blaNDM-1 and blaKPC-2 gene. Correspondingly, the mcr-1 gene is being reported from Pakistan [115]. Further molecular studies about the strain type, sequence type, and plasmid typing are required to better understand the resistant magnitude. We also suggest the clinicians for appropriate colistin and carbapenem prescriptions, as bacteria developed plasmid-mediated resistance against them having the horizontal transferability [116]. Several gaps in the surveillance were noted, i-e, we did not find any study from Baluchistan province. However, most of the studies are from Karachi, especially from Agha khan university hospital, which receives samples via its collection points in 190 major cities and towns across the country [62]. 82.79, 6.5, 33.3, 28.8, and 18.3% of data were not available for the source of infection, date of sample collection, demography, patient type, and susceptibility testing standard, respectively. Such gaps make their data suspicious, and we encouraged the researcher to address all these gaps in their future studies. Along with that, more research work is required from Baluchistan province and small cities and towns of Pakistan. The molecular studies required a distinctive focus to combat this pan-drug resistant phenomenon.
The present study focuses on antibiotic resistance, specifically in Pakistan; however, their implication is worldwide. Pakistan has a strategically important geographical location as an adjacent neighbor of the Middle East with a shared border with China, Afghanistan, Iran, India, and less than the one-kilometer distance from Uzbekistan (central Asian state) [117]. It is known that resistant species from its reservoir can spread to other regions of the world via human, water, and animals [118]. In the case of Pakistan, its consequences seem the most significant threat.
Our study’s limitation is that we do not have data from Baluchistan province; also, most of the studies are from the capitals of provinces, which might not be an appropriate depiction of the whole country. In 83.9% of studies, the infection sources were not determined as usually, the hospital-acquired pathogens are more resistant. We find out the MR of at least two studies because of more isolates in each study. Furthermore, different kinds of data about patient type, demographic, and methodologies are combined. However, our study shows an exclusive preview of antibiotic-resistance in Pakistan. Researchers must follow all the gaps in their future studies.
On a vaster glimpse, the antibiotic resistance in Pakistan is very high; both the community and health care seating must need special attention to this issue. For the community, the awareness is required about the cautious use and completion of dosage. Self-medication must be prohibited among the community. Guidelines of antibiotic practice in husbandry and human wellbeing should be practical, founded on Pakistan’s antimicrobial resistance network (PARN) to lessen the hazard of alarming antimicrobial resistance. Transmission of antibiotic-resistant bacteria in health care amenities can be reduced by adopting recommended precautionary measures such as contact precautions, personal hand cleanliness, educating, training healthcare workers, and lessening devices’ use.
Conclusions
The present study summarizes the surveillance data of antibiotic resistance from Pakistan and emphasizes the four significant outcomes. 1) The prevalence of AMR to commonly prescribed antibiotics is very high in Pakistan. 2) Substantial gaps in surveillance are found i-e no study about antibiotic resistance was reported for Baluchistan province. Also, the number of studies for certain bacteria was too insufficient to calculate their resistance patterns. 3) Gaps in information for methodological data are noted in several studies, making their quality suspicious and difficult for analysis. 4) Only a few molecular studies are available which are required for effective and apposite use of therapeutic agents. Therefore, there is a necessity for regularization of surveillance practice and continuous regional and nationwide surveillance, molecular studies, along with specific actions to combat the hazard associated with the increase of AMR.
Abbreviations
- aac(6′)- Ib-cr :
-
Aminoglycoside 6′-N-acetyl transferase type Ib-cr (Citrobacter freundii)
- aadA1 :
-
Aminoglycoside adenyltransferase-A1 gene
- ABR:
-
Antibiotic-resistant
- AMP:
-
Ampicillinase, β-lactamase of ESBL-M type
- AMR:
-
Antimicrobial resistance
- bla :
-
Gene encoding β-lactamase
- blt :
-
Efflux transporter (promiscuous, acetylated polyamines efflux) (Bacillus Subtilis)
- BSAC:
-
British Society for Antimicrobial Chemotherapy
- BSI:
-
Blood stream infection
- catA1 :
-
Chloramphenicol acetyl transferase
- catP :
-
Chloramphenicol O-acetyltransferase gene
- cfr :
-
23S rRNA (adenine(2503)-C(8))-methyltransferase gene
- CLSI:
-
Clinical & Laboratory Standards Institute
- CTXM:
-
Cefotaximase Munich, β-lactamase of ESBL-A type
- DDM:
-
Disk Diffusion Method
- dfrA7 :
-
Dihydrofolate reductase-A7 gene
- E Test:
-
Epsilometer test
- E. coli :
-
Escherichia coli
- ESBL:
-
Extended Spectrum Beta-Lactamase
- EUCAST:
-
European Committee on Antibiotic Susceptibility Testing
- gyrA:
-
DNA gyrase subunit A
- H. pylori :
-
Helicobacter Pylori
- CI:
-
Confidence Interval
- KPC-2:
-
Klebsiella pneumoniae carbapenemase-2
- MBL:
-
Metallo-beta-lactamase
- MDR:
-
Multidrug-Resistance
- MSSA:
-
Methicillin-susceptible Staphylococcus aureus
- MR:
-
Median Resistance
- MRSA:
-
Methicillin-resistant Staphylococcus aureus
- NDM-1:
-
New Delhi metallo-beta-lactamase-1
- OXA:
-
Oxacillinase-type β –lactamase
- PARN:
-
Pakistan’s antimicrobial resistance network
- qepA :
-
Quinolone pump gene
- qnr :
-
Quinolone-resistance gene
- S. aureus :
-
Staphylococcus aureus
- SHV:
-
Sulfhydryl variable, β -lactamase of ESBL-A type
- StrA/B :
-
Streptomycin phosphotransferase-A/B
- sul2 :
-
Sulfonamide-resistant dihydropteroate synthase
- TEM:
-
Temoneira, β -lactamase of ESBL-A type
- tetA :
-
Tetracycline efflux MFS transporter-A gene
- tetB :
-
Tetracycline efflux MFS transporter-B gene
- VanA :
-
Vanillate O-demethylase oxygenase subunit (4-hydroxy-3-methoxybenzoate demethylase)
- VRE:
-
Vancomycin-resistant Enterococcus
- VRSA:
-
Vancomycin resistant Staphylococcus aureus
- XDR:
-
Extensive Drug Resistance
References
Boolchandani M, D’Souza AW, Dantas G. Sequencing-based methods and resources to study antimicrobial resistance. Nat Rev Gen. 2019:1.
Padiyara P, Inoue H, Sprenger M. Global governance mechanisms to address antimicrobial resistance. Infect Dis (Auckl). 2018;11:1178633718767887.
Kang CI, Song JH. Antimicrobial resistance in Asia: current epidemiology and clinical implications. Infect Chemother. 2013;45(1):22–31.
Abrar S, Hussain S, Khan RA, Ain NU, Haider H, Riaz S. Prevalence of extended-spectrum-β-lactamase-producing Enterobacteriaceae: first systematic meta-analysis report from Pakistan. Antimicrobial Resist Infect Control. 2018;7(1):26.
Yasmin F, Akhtar N, Hameed A. In vitro synergistic effect of ciprofloxacin with aminoglycosides against multidrug resistant-Pseudomonas aeruginosa. Pak J Pharm Sci. 2013;26(5):1041–4 https://pubmed.ncbi.nlm.nih.gov/24035966/.
Qamar FN, Yousafzai MT, Khalid M, Kazi AM, Lohana H, Karim S, Khan A, Hotwani A, Qureshi S, Kabir F. Outbreak investigation of ceftriaxone-resistant Salmonella enterica serotype Typhi and its risk factors among the general population inS Hyderabad, Pakistan: a matched case-control study. Lancet Infect Dis. 2018;18(12):1368–76.
Latif S, Anwar MS, Ahmad I. Bacterial pathogens responsible for blood stream infection (BSI) and pattern of drug resistance in a tertiary care hospital of Lahore. Biomedica. 2009;25(2):101–5 https://pdfs.semanticscholar.org/e089/45412f8d65ab59660d27f26cb9a9bb7cd9fd.pdf.
Kaleem F, Usman J, Hassan A, Khan A. Frequency and susceptibility pattern of metallo-beta-lactamase producers in a hospital in Pakistan. J Infect Develop Countries. 2010;4(12):810–3.
Pesesky MW, Hussain T, Wallace M, Wang B, Andleeb S, Burnham CA, Dantas G. KPC and NDM-1 genes in related Enterobacteriaceae strains and plasmids from Pakistan and the United States. Emerg Infect Dis. 2015;21(6):1034–7.
Hameed F, Khan MA, Muhammad H, Sarwar T, Bilal H, Rehman TU. Plasmid-mediated mcr-1 gene in Acinetobacter baumannii and Pseudomonas aeruginosa: first report from Pakistan. Rev Soc Bras Med Trop. 2019;52:e20190237.
Faizullah M, Nisar Ur R, Umar M, Anwar M, Sarfraz M. A cross-sectional study on knowledge, attitude and practices of medical doctors towards antibiotic prescribing patterns and resistance in Khyber Pakhtun Khawah, Pakistan. J Appl Pharmaceut Sci. 2017;7:38–46. https://doi.org/10.7324/JAPS.2017.71205.
Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–27.
Sohail M, Rashid A, Aslam B, Waseem M, Shahid M, Akram M, Khurshid M, Rasool MH. Antimicrobial susceptibility of Acinetobacter clinical isolates and emerging antibiogram trends for nosocomial infection management. Rev Soc Bras Med Trop. 2016;49(3):300–4.
Uddin F, McHugh TD, Roulston K, Platt G, Khan TA, Sohail M. Detection of carbapenemases, AmpC and ESBL genes in Acinetobacter isolates from ICUs by DNA microarray. J Microbiol Methods. 2018;155:19–23.
Khurshid M, Rasool MH, Ashfaq UA, Aslam B, Waseem M, Xu Q, Zhang X, Guo Q, Wang M. Dissemination of blaOXA-23 harboring Carbapenem-Resistant Acinetobacter baumannii clones in Pakistan. J Glob Antimicrob Resist. 2020;21:357–62.
Gilani M, Latif M, Gilani M, Saad N, Ansari M, Gilani M, Waseem H, Naeem A. Efficacy of antimicrobials against multidrug-resistant acinetobacter baumannii from patients in a Tertiary Care Hospital. Microb Drug Resist. 2020;26(6):681–4.
Hasan B, Perveen K, Olsen B, Zahra R. Emergence of carbapenem-resistant Acinetobacter baumannii in hospitals in Pakistan. J Med Microbiol. 2014;63(Pt 1):50–5.
Khurshid M, Rasool MH, Ashfaq UA, Aslam B, Waseem M. Emergence of ISAba1 harboring carbapenem-resistant Acinetobacter baumannii isolates in Pakistan. Future Microbiol. 2017;12:1261–9.
Ain NU, Iftikhar A, Bukhari SS, Abrar S, Hussain S, Haider MH, Rasheed F, Riaz S. High frequency and molecular epidemiology of metallo-beta-lactamase-producing gram-negative bacilli in a tertiary care hospital in Lahore, Pakistan. Antimicrob Resist Infect Control. 2018;7:128.
Shakoor S, Khan E, Zafar A, Hasan R. In vitro activity of tigecycline and other tetracyclines against carbapenem-resistant Acinetobacter species: report from a tertiary care Centre in Karachi, Pakistan. Chemotherapy. 2010;56(3):184–9.
Irfan S, Turton JF, Mehraj J, Siddiqui SZ, Haider S, Zafar A, Memon B, Afzal O, Hasan R. Molecular and epidemiological characterisation of clinical isolates of carbapenem-resistant Acinetobacter baumannii from public and private sector intensive care units in Karachi, Pakistan. J Hosp Infect. 2011;78(2):143–8.
Karah N, Khalid F, Wai SN, Uhlin BE, Ahmad I. Molecular epidemiology and antimicrobial resistance features of Acinetobacter baumannii clinical isolates from Pakistan. Ann Clin Microbiol Antimicrob. 2020;19(1):2.
Khurshid M, Rasool MH, Siddique MH, Azeem F, Naeem M, Sohail M, Sarfraz M, Saqalein M, Taj Z, Nisar MA, et al. Molecular mechanisms of antibiotic co-resistance among carbapenem resistant Acinetobacter baumannii. J Infect Dev Ctries. 2019;13(10):899–905.
Saeed MA, Haque A, Ali A, Mohsin M, Bashir S, Tariq A, Afzal A, Iftikhar T, Sarwar Y. A profile of drug resistance genes and integrons in E. coli causing surgical wound infections in the Faisalabad region of Pakistan. J Antibiot (Tokyo). 2009;62(6):319–23.
Ali I, Kumar N, Ahmed S, Dasti JI: Antibiotic resistance in uropathogenic E. coli strains isolated from non-hospitalized patients in pakistan. J Clin Diagn Res 2014, 8(9):Dc01–Dc04. https://doi: https://doi.org/10.7860/jcdr/2014/7881.4813.
Ali I, Shabbir M. Antibiotics susceptibility patterns of uropathogenic E. coli with special reference to fluoroquinolones in different age and gender groups. J Pak Med Assoc. 2017;67(8):1161–5 https://pubmed.ncbi.nlm.nih.gov/28839298/.
Hussain Gilani SY, Ali Shah SR, Ahmad N, Bibi S. Antimicrobial resistance patterns in community acquired urinary tract infections. J Ayub Med Coll Abbottabad. 2016;28(3):572–4 https://pubmed.ncbi.nlm.nih.gov/28712238.
Kumar N, Nahid F, Zahra R. Association of virulence factors, phylogenetic groups and antimicrobial resistance markers in Escherichia coli from Badin city, Pakistan. J Chemother. 2017;29(1):8–13.
Parveen A, Sultan F, Raza A, Zafar W, Nizamuddin S, Mahboob A, Saleem S, Nazeer SH. Bacteraemia caused by Escherichia coli in cancer patients at a specialist center in Pakistan. J Pak Med Assoc. 2015;65(12):1271–6 https://www.pakmedinet.com/24676.
Nazir H, Cao S, Hasan F, Hughes D. Can phylogenetic type predict resistance development? J Antimicrob Chemother. 2011;66(4):778–87.
Younas M, Siddiqui F, Noreen Z, Bokhari SS, Gomez-Duarte OG, Wren BW, Bokhari H. Characterization of enteropathogenic Escherichia coli of clinical origin from the pediatric population in Pakistan. Trans R Soc Trop Med Hyg. 2016;110(7):414–20.
Abrar S, Ain NU, Liaqat H, Hussain S, Rasheed F, Riaz S. Distribution of bla (CTX - M) , bla (TEM) , bla (SHV) and bla (OXA) genes in Extended-spectrum-β-lactamase-producing Clinical isolates: A three-year multi-center study from Lahore, Pakistan. Antimicrob Resist Infect Control. 2019;8:80.
Abrar S, Vajeeha A, Ul-Ain N, Riaz S. Distribution of CTX-M group I and group III beta-lactamases produced by Escherichia coli and klebsiella pneumoniae in Lahore, Pakistan. Microb Pathog. 2017;103:8–12.
Farooq L, Ahmed SN, Khan MAU, Ali A, Mehmood S, Arif H. In vitro activity of Ceftolozane/Tazobactam for the treatment of complicated urinary tract infections by Escherichia coli in the era of antibiotic resistance “rejuvenate the mystery”. J Pharmaceut Res Int. 2019:1–7.
Sabir S, Anjum A, Ijaz T, Ali M, Khan M-U-R, Nawaz M: Isolation and antibiotic susceptibility of E. coli from urinary tract infections in a tertiary care hospital. Pak J Med Sci 2014, 30:389–392. https://doi: https://doi.org/10.12669/pjms.302.4289.
Jameel N-U-A, Ejaz H, Zafar A, Amin H: Multidrug resistant AmpC β-lactamase producing Escherichia coli isolated from a paediatric hospital. Pak J Med Sci 2014, 30:181–184. https://doi: https://doi.org/10.12669/pjms.301.4045
Habeeb MA, Haque A, Iversen A, Giske CG. Occurrence of virulence genes, 16S rRNA methylases, and plasmid-mediated quinolone resistance genes in CTX-M-producing Escherichia coli from Pakistan. Eur J Clin Microbiol Infect Dis. 2014;33(3):399–409.
Khalil U, Younus M, Asghar N, Siddiqui F, Gomez-Duarte OG, Wren BW, Bokhari H. Phenotypic and genotypic characterization of enteroaggregative Escherichia coli isolates from pediatric population in Pakistan. Apmis. 2016;124(10):872–80.
Fatima S, Muhammad IN, Jamil S, Siddiqui T, Khatoon H, Usman S. Prevalence of CTX-M variants in ESBL producing multidrug-resistant Enterobacteriaceae from outpatients in Karachi, Pakistan. Latin Am J Pharm. 2019;38(8):1516–21 http://www.latamjpharm.org/resumenes/38/8/LAJOP_38_8_1_5.pdf.
Perry JD, Naqvi SH, Mirza IA, Alizai SA, Hussain A, Ghirardi S, Orenga S, Wilkinson K, Woodford N, Zhang J, et al. Prevalence of faecal carriage of Enterobacteriaceae with NDM-1 carbapenemase at military hospitals in Pakistan, and evaluation of two chromogenic media. J Antimicrob Chemother. 2011;66(10):2288–94.
Jamil J, Haroon M, Sultan A, Khan MA, Gul N. Prevalence, antibiotic sensitivity and phenotypic screening of ESBL/MBL producer E. coli strains isolated from urine; district Swabi, KP, Pakistan. J Pak Med Assoc. 2018;68(11):1704–7 https://jpma.org.pk/article-details/8941.
Rana F, Siddiqui S, Khan A, Siddiqui F, Noreen Z, Bokhari S, Bokhari H. Resistance patterns of diversified phylogroups of Escherichia coli associated with mothers having history of preterm births in Pakistan. J Matern Fetal Neonatal Med. 2017;30(1):68–73.
Qamar S, Durrani MA, Raoof S. Resistant pattern of enterobacteriaceae isolates from surgical wards of tertiary care hospital. Pak J Pharmacol. 2010;27(1):37–41 https://gg.gds.workers.dev/scholar?cluster=12967515562020741083&hl=en&as_sdt=0,5.
Rasheed F, Campbell BJ, Alfizah H, Varro A, Zahra R, Yamaoka Y, Pritchard DM. Analysis of clinical isolates of helicobacter pylori in Pakistan reveals high degrees of pathogenicity and high frequencies of antibiotic resistance. Helicobacter. 2014;19(5):387–99.
Khan A, Farooqui A, Manzoor H, Akhtar SS, Quraishy MS, Kazmi SU. Antibiotic resistance and cagA gene correlation: a looming crisis of helicobacter pylori. World J Gastroenterol. 2012;18(18):2245–52.
Siddiqui TR, Ahmed W, Arif A, Bibi S, Khan A. Emerging trends of antimicrobial resistance in helicobacter pylori isolates obtained from Pakistani patients: the need for consideration of amoxicillin and clarithromycin. J Pak Med Assoc. 2016;66(6):710–6 https://pakmedinet.com/26252.
Younas S, Ejaz H, Zafar A, Ejaz A, Saleem R, Javed H. AmpC beta-lactamases in Klebsiella pneumoniae: an emerging threat to the paediatric patients. J Pak Med Assoc. 2018;68(6):893–7 https://pubmed.ncbi.nlm.nih.gov/30325907/.
Qamar MU, Walsh TR, Toleman MA, Tyrrell JM, Saleem S, Aboklaish A, Jahan S. Dissemination of genetically diverse NDM-1, −5, −7 producing-gram-negative pathogens isolated from pediatric patients in Pakistan. Future Microbiol. 2019;14:691–704.
Zafar S, Hanif S, Akhtar H, Faryal R. Emergence of hypervirulent K pneumoniae causing complicated UTI in kidney stone patients. Microb Pathog. 2019;135:103647.
Humayun A, Siddiqui FM, Akram N, Saleem S, Ali A, Iqbal T, Kumar A, Kamran R, Bokhari H. Incidence of metallo-beta-lactamase-producing Klebsiella pneumoniae isolates from hospital setting in Pakistan. Int Microbiol. 2018;21(1–2):73–8.
Khan E, Ejaz M, Zafar A, Jabeen K, Shakoor S, Inayat R, Hasan R. Increased isolation of ESBL producing Klebsiella pneumoniae with emergence of carbapenem resistant isolates in Pakistan: report from a tertiary care hospital. J Pak Med Assoc. 2010;60(3):186–90 https://pubmed.ncbi.nlm.nih.gov/20225774/.
Javed H, Ejaz H, Zafar A, Rathore AW. Metallo-beta-lactamase producing Escherichia coli and Klebsiella pneumoniae: a rising threat for hospitalized children. J Pak Med Assoc. 2016;66(9):1068–72 https://pubmed.ncbi.nlm.nih.gov/27654721/.
Mal PB, Jabeen K, Farooqi J, Unemo M, Khan E. Antimicrobial susceptibility testing of Neisseria gonorrhoeae isolates in Pakistan by Etest compared to calibrated dichotomous sensitivity and clinical laboratory standards institute disc diffusion techniques. BMC Microbiol. 2016;16(1):236.
Jabeen K, Nizamuddin S, Irfan S, Khan E, Malik F, Zafar A. Increasing Trend of Resistance to Penicillin, Tetracycline, and Fluoroquinolone Resistance in Neisseria gonorrhoeae from Pakistan (1992-2009). J Trop Med. 2011:960501.
Ullah F, Malik SA, Ahmed J. Antimicrobial susceptibility and ESBL prevalence in Pseudomonas aeruginosa isolated from burn patients in the north west of Pakistan. Burns. 2009;35(7):1020–5.
Sattar H, Toleman M, Nahid F, Zahra R. Co-existence of blaNDM-1 and blaKPC-2 in clinical isolates of Klebsiella pneumoniae from Pakistan. J Chemother. 2016;28(4):346–9.
Farooq L, Memon Z, Ismail MO, Sadiq S. Frequency and antibiogram of multi-drug resistant pseudomonas aeruginosa in a tertiary Care Hospital of Pakistan. Pak J Med Sci. 2019;35(6):1622–6.
Ullah W, Qasim M, Rahman H, Bari F, Khan S, Rehman ZU, Khan Z, Dworeck T, Muhammad N. Multi drug resistant Pseudomonas aeruginosa: pathogen burden and associated antibiogram in a tertiary care hospital of Pakistan. Microb Pathog. 2016;97:209–12.
Qamar MU, Nahid F, Walsh TR, Kamran R, Zahra R. Prevalence and clinical burden of NDM-1 positive infections in pediatric and neonatal patients in Pakistan. Pediatr Infect Dis J. 2015;34(4):452–4.
Ahmad M, Hassan M, Khalid A, Tariq I, Asad MH, Samad A, Mahmood Q, Murtaza G. Prevalence of extended Spectrum beta-lactamase and antimicrobial susceptibility pattern of clinical isolates of Pseudomonas from patients of Khyber Pakhtunkhwa, Pakistan. Biomed Res Int. 2016:6068429.
Qamar FN, Yousafzai MT, Sultana S, Baig A, Shakoor S, Hirani F, Wassay A, Khushboo S, Mehmood J, Freeman A et al: A Retrospective Study of Laboratory-Based Enteric Fever Surveillance, Pakistan, 2012-2014. J Infect Dis 2018, 218(suppl_4):S201-s205. https://doi: https://doi.org/10.1093/infdis/jiy205
Qamar FN, Azmatullah A, Kazi AM, Khan E, Zaidi AK. A three-year review of antimicrobial resistance of Salmonella enterica serovars Typhi and Paratyphi a in Pakistan. J Infect Dev Ctries. 2014;8(8):981–6.
Yousafzai MT, Qamar FN, Shakoor S, Saleem K, Lohana H, Karim S, Hotwani A, Qureshi S, Masood N, Rauf M, et al. Ceftriaxone-resistant Salmonella Typhi outbreak in Hyderabad City of Sindh, Pakistan: high time for the introduction of typhoid conjugate vaccine. Clin Infect Dis. 2019;68(Suppl 1):S16–s21.
Klemm EJ, Shakoor S, Page AJ, Qamar FN, Judge K, Saeed DK, Wong VK, Dallman TJ, Nair S, Baker S, et al. Emergence of an extensively drug-resistant Salmonella enterica Serovar Typhi Clone Harboring a Promiscuous Plasmid Encoding Resistance to Fluoroquinolones and Third-Generation Cephalosporins. mBio. 2018;9(1):e00105–18.
Aziz S, Malik L. Emergence of multi-resistant enteric infection in a Paediatric unit of Karachi, Pakistan. J Pak Med Assoc. 2018;68(12):1848–50 https://www.pakmedinet.com/35367.
Khan MI, Soofi SB, Ochiai RL, Khan MJ, Sahito SM, Habib MA, Puri MK, Von Seidlein L, Park JK, You YA, et al. Epidemiology, clinical presentation, and patterns of drug resistance of Salmonella Typhi in Karachi, Pakistan. J Infect Dev Ctries. 2012;6(10):704–14.
Qaiser S, Irfan S, Khan E, Ahsan T, Zafar A. In vitro susceptibility of typhoidal salmonellae against newer antimicrobial agents: a search for alternate treatment options. J Pak Med Assoc. 2011;61(5):462–5 https://pubmed.ncbi.nlm.nih.gov/22204180/.
Afzal A, Sarwar Y, Ali A, Maqbool A, Salman M, Habeeb MA, Haque A. Molecular evaluation of drug resistance in clinical isolates of Salmonella enterica serovar Typhi from Pakistan. J Infect Dev Ctries. 2013;7(12):929–40.
Hussain A, Satti L, Hanif F, Zehra NM, Nadeem S, Bangash TM, Peter A. Typhoidal Salmonella strains in Pakistan: an impending threat of extensively drug-resistant Salmonella Typhi. Eur J Clin Microbiol Infect Dis. 2019;38(11):2145–9.
Zafar A, Hasan R, Nizami SQ, von Seidlein L, Soofi S, Ahsan T, Chandio S, Habib A, Bhutto N, Siddiqui FJ, et al. Frequency of isolation of various subtypes and antimicrobial resistance of Shigella from urban slums of Karachi, Pakistan. Int J Infect Dis. 2009;13(6):668–72.
Tariq A, Haque A, Ali A, Bashir S, Habeeb MA, Salman M, Sarwar Y. Molecular profiling of antimicrobial resistance and integron association of multidrug-resistant clinical isolates of Shigella species from Faisalabad, Pakistan. Can J Microbiol. 2012;58(9):1047–54.
Raza A, Mahboob A, Nizammudin S, Nazeer SH, Sultan F. Shigella infections: a two year experience in cancer patients. J Pak Med Assoc. 2016;66(1):37–9 https://www.jpma.org.pk/article-details/7580.
Khan E, Jabeen K, Ejaz M, Siddiqui J, Shezad MF, Zafar A. Trends in antimicrobial resistance in Shigella species in Karachi, Pakistan. J Infect Dev Ctries. 2009;3(10):798–802.
Fasih N, Zafar A, Khan E, Jabeen K, Hasan R. Clonal dissemination of vanA positive Enterococcus species in tertiary care hospitals in Karachi, Pakistan. J Pak Med Assoc. 2010;60(10):805–9 https://pubmed.ncbi.nlm.nih.gov/21381606/.
Yameen MA, Iram S, Mannan A, Khan SA, Akhtar N. Nasal and perirectal colonization of vancomycin sensitive and resistant enterococci in patients of paediatrics ICU (PICU) of tertiary health care facilities. BMC Infect Dis. 2013;13:156.
Hussain A, Sohail M, Abbas Z. Prevalence of Enterococcus faecalis mediated UTI and its current antimicrobial susceptibility pattern in Lahore, Pakistan. J Pak Med Assoc. 2016;66(10):1232–6 https://pubmed.ncbi.nlm.nih.gov/27686295/.
Rasool MS, Siddiqui F, Ajaz M, Rasool SA, Hafiz S. Antibiotic resistance trends in indigenous methicillin resistant Staphylococcus aureus (MRSA) associated with bacteremia. Pak J Pharmacol. 2017;34(1 & 2):39–44 http://www.pakjp.pk/wp-content/uploads/pdfs/34/1/5-PJP-027.pdf.
Idrees F, Jabeen K, Khan MS, Zafar A. Antimicrobial resistance profile of methicillin resistant staphylococcal aureus from skin and soft tissue isolates. J Pak Med Assoc. 2009;59(5):266–9 https://www.pakmedinet.com/13971.
Younus A, Faiz M, Saleem M, Saghir S. Antimicrobial susceptibility patterns and characterization of clinical isolates of Staphylococcus aureus in immunocompromised cancer patients undergoing chemotherapy in Pakistan. J Chemother. 2009;21(1):31–5.
Azhar A, Rasool S, Haque A, Shan S, Saeed M, Ehsan B, Haque A. Detection of high levels of resistance to linezolid and vancomycin in Staphylococcus aureus. J Med Microbiol. 2017;66(9):1328–31.
Rafiq MS, Rafiq MI, Khan T, Rafiq M, Khan MM. Effectiveness of simple control measures on methicillin-resistant Staphylococcus aureus infection status and characteristics with susceptibility patterns in a teaching hospital in Peshawar. J Pak Med Assoc. 2015;65(9):915–20 https://jpma.org.pk/article-details/7450?article_id=7450.
Hanif E, Hassan SA: Evaluation of antibiotic resistance pattern in clinical isolates of Staphylococcus aureus. Pak J Pharm Sci 2019, 32(4(Supplementary)):1749–1753. https://pubmed.ncbi.nlm.nih.gov/31680068/.
Nizamuddin S, Irfan S, Zafar A. Evaluation of prevalence of low and high level mupirocin resistance in methicillin resistant Staphylococcus aureus isolates at a tertiary care hospital. J Pak Med Assoc. 2011;61(6):519–21 https://pubmed.ncbi.nlm.nih.gov/22204200/.
Ehsan MM, Memon Z, Ismail MO, Fatima G. Identification and antibiotic susceptibility pattern of coagulase-negative staphylococci in various clinical specimens. Pak J Med Sci. 2013;29(6):1420–4.
Pervaz S. Isolation of methicillin-resistant Staphylococcus aureus from burn centers and evaluation of antimicrobial efficacy, in Faisalabad, Pakistan. Sci J Med Res. 2018;02:158–65.
Sohail M, Latif Z. Molecular analysis, biofilm formation, and susceptibility of methicillin-resistant Staphylococcus aureus strains causing community- and health care-associated infections in central venous catheters. Rev Soc Bras Med Trop. 2018;51(5):603–9.
Kaleem F, Usman J, Hassan A, Omair M, Khalid A, Uddin R. Sensitivity pattern of methicillin resistant Staphylococcus aureus isolated from patients admitted in a tertiary care hospital of Pakistan. Iran J Microbiol. 2010;2(3):143–6 https://pubmed.ncbi.nlm.nih.gov/22347563/.
Ahmed A, Hussain S, Ijaz T, Hashemy I. Susceptibility of methicillin-resistant Staphylococcus aureus and enterococci to teicoplanin in Pakistan: the MRSET study. J Pak Med Assoc. 2014;64(3):256–9 https://jpma.org.pk/article-details/6110?article_id=6110.
Saleem F, Fasih N, Zafar A. Susceptibility pattern of methicillin resistant Staphylococcus aureus to vancomycin and other alternate agents: report from a private sector hospital laboratory. J Pak Med Assoc. 2017;67(11):1743–6 https://ecommons.aku.edu/pakistan_fhs_mc_pathol_microbiol/697/.
Shah AA, Jamil B, Naseem S, Khan AW, Ali Y, Hussain K, Abbasi SA. Susceptibility pattern of tracheal tube isolates from intensive care unit of Fauji Foundation hospital Rawalpindi. J Pak Med Assoc. 2019;69(7):981–4 https://pubmed.ncbi.nlm.nih.gov/31308567/.
Ullah O, Khan A, Ambreen A, Ahmad I, Akhtar T, Gandapor AJ, Khan AM: Antibiotic Sensitivity pattern of Bacterial Isolates of Neonatal Septicemia in Peshawar, Pakistan. Arch Iran Med 2016, 19(12):866–869. https://doi: 0161912/aim.009.
Hubab M, Ullah O, Hayat A, Rehman MU, Sultana N. Antibiotic susceptibility profile of bacterial isolates from post-surgical wounds of patients in tertiary care hospitals of Peshawar, Pakistan. J Pak Med Assoc. 2018;68(10):1517–21 https://jpma.org.pk/article-details/8896?article_id=8896.
Ali AM. Bacteriological Spectrum of UTI and their Antibiograms in a tertiary care cardiac hospital, Rawalpindi, Pakistan. EC Cardiol. 2019;6:766–71 https://www.ecronicon.com/eccy/pdf/ECCY-06-00359.pdf.
Sohail M, Khurshid M, Saleem HG, Javed H, Khan AA. Characteristics and antibiotic resistance of urinary tract pathogens isolated from Punjab, Pakistan. Jundishapur J Microbiol. 2015;8(7):e19272.
Sabir R, Alvi SFD, Fawwad A, Basit A. Antibiogram of Pseudomonas aeruginosa and methicillin-resistant Staphylococcus aureus in patients with diabetes. Pak J Med Sci. 2014;30(4):814–8.
Ali I, Butt M: Antibiotic susceptibility pattern of bacterial isolates from patients of respiratory tract infection at 43 centers in Punjab, Pakistan, Clin Exp Pharmacol 2017, 7(229):2161–1459.1000229. https://doi.org/https://doi.org/10.4172/2161-1459.1000229.
Amjad SS, Zafar J, Shams N. Bacteriology of diabetic foot in tertiary care hospital; frequency, antibiotic susceptibility and risk factors. J Ayub Med Coll Abbottabad. 2017;29(2):234–40 https://www.pakmedinet.com/29788.
Fayyaz I, Rasheed MA, Ashraf M, Bukhsh A, Wadood A. Determination of bacterial etiological agents, sensitivity pattern and clinical outcome of patients with bacterial endocarditis at Punjab Institute of Cardiology, Lahore. J Pak Med Assoc. 2014;64(12):1384–8 https://pubmed.ncbi.nlm.nih.gov/25842583/.
Mir F, Tikmani SS, Shakoor S, Warraich HJ, Sultana S, Ali SA, Zaidi AK. Incidence and etiology of omphalitis in Pakistan: a community-based cohort study. J Infect Dev Ctries. 2011;5(12):828–33.
Miyan Z, Fawwad A, Sabir R, Basit A. Microbiological pattern of diabetic foot infections at a tertiary care center in a developing country. J Pak Med Assoc. 2017;67(5):665–9 https://pubmed.ncbi.nlm.nih.gov/28507348/.
Zafar A, Hasan R, Nizamuddin S, Mahmood N, Mukhtar S, Ali F, Morrissey I, Barker K, Torumkuney D. Antibiotic susceptibility in Streptococcus pneumoniae, Haemophilus influenzae and Streptococcus pyogenes in Pakistan: a review of results from the survey of antibiotic resistance (SOAR) 2002-15. J Antimicrob Chemother. 2016;71(Suppl 1):i103–9.
Furqan S, Paracha SA. Frequency of Streptococcus pneumonia and Haemophilus influenza in acute exacerbation of chronic obstructive airway disease and their sensitivity to levofloxacin. J Pak Med Assoc. 2014;64(4):399–402 https://pubmed.ncbi.nlm.nih.gov/22347563/.
Ahmed I, Rabbi MB, Sultana S. Antibiotic resistance in Bangladesh: a systematic review. Int J Infect Dis. 2019;80:54–61.
Tadesse BT, Ashley EA, Ongarello S, Havumaki J, Wijegoonewardena M, Gonzalez IJ, Dittrich S. Antimicrobial resistance in Africa: a systematic review. BMC Infect Dis. 2017;17(1):616.
Mahindroo J, Thanh DP, Nguyen TNT, Mohan B, Thakur S, Baker S, Taneja N. Endemic fluoroquinolone-resistant Salmonellaenterica serovar Kentucky ST198 in northern India. Microb Genom. 2019;5(7):e000275.
Shankar PR, Balasubramanium R. Antimicrobial resistance: global report on surveillance. Aust Med J. 2014;7:237 http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf.
Shafquat Y, Jabeen K, Farooqi J, Mehmood K, Irfan S, Hasan R, Zafar A. Antimicrobial susceptibility against metronidazole and carbapenem in clinical anaerobic isolates from Pakistan. Antimicrob Resist Infect Control. 2019;8:99.
Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643.
Kourtis AP, Hatfield K, Baggs J, Mu Y, See I, Epson E, Nadle J, Kainer MA, Dumyati G, Petit S, et al. Vital signs: epidemiology and recent trends in methicillin-resistant and in methicillin-susceptible Staphylococcus aureus bloodstream infections - United States. MMWR Morb Mortal Wkly Rep. 2019;68(9):214–9.
Albrecht VS, Limbago BM, Moran GJ, Krishnadasan A, Gorwitz RJ, McDougal LK, Talan DA. Group EMINS: Staphylococcus aureus colonization and strain type at various body sites among patients with a closed abscess and uninfected controls at U.S. emergency departments. J Clin Microbiol. 2015;53(11):3478–84.
Markwart R, Willrich N, Haller S, Noll I, Koppe U, Werner G, Eckmanns T, Reuss A. The rise in vancomycin-resistant Enterococcus faecium in Germany: data from the German antimicrobial resistance surveillance (ARS). Antimicrob Resist Infect Control. 2019;8:147.
Orsi GB, Ciorba V. Vancomycin resistant enterococci healthcare associated infections. Ann Ig. 2013;25(6):485–92. https://doi.org/10.7416/ai.2013.1948.
Hadjadj L, Syed MA, Bushra J, Abbasi SA, Rolain JM. Emergence of Vancomycin-resistant Enterococcus faecium ST 80 in Pakistan. Surg Infect. 2019;20(6):524–5.
Munita JM, Arias CA. Mechanisms of Antibiotic Resistance. Microbiol Spectr. 2016;4(2).
Bilal H, Hameed F, Khan MA, Khan S, Yang X, Rehman TU. Detection of mcr-1 gene in extended-Spectrum β-lactamase-producing Klebsiella pneumoniae from human urine samples in Pakistan. Jundishapur J Microbiol. 2020;13(4):e96646.
Martino F, Tijet N, Melano R, Petroni A, Heinz E, De Belder D, Faccone D, Rapoport M, Biondi E, Rodrigo V, et al. Isolation of five Enterobacteriaceae species harbouring blaNDM-1 and mcr-1 plasmids from a single paediatric patient. PLoS One. 2019;14(9):e0221960.
Pakistan. Department of State publication Background notes series 1987:1–10. https://www.worldcat.org/title/background-notes-pakistan/oclc/10962212
Sjölund M, Bonnedahl J, Hernandez J, Bengtsson S, Cederbrant G, Pinhassi J, Kahlmeter G, Olsen B. Dissemination of multidrug-resistant bacteria into the Arctic. Emerg Infect Dis. 2008;14(1):70–2.
Acknowledgments
The authors are thankful to Dr. Akhtar Nadhman from the Institute of Integrative Biosciences Faculty, CECOS University Hayatabad, Peshawar, to analyze the study design and manuscript.
Availability of data
Data supporting our findings can be found through the corresponding author (email: xingyuan@ahu.edu.cn) or the senior author (email: bilal.microbiologist@yahoo.com).
Funding
This work was supported by grants from Natural Science Foundation of China (number 31771310 to Xingyuan Yang) and Anhui Province Natural Science Foundation (number 1708085MC67 to Xingyuan Yang). The funders had no rule in the study design, data collection and analysis, manuscript writing, proofreading, and decision to publish.
Author information
Authors and Affiliations
Contributions
Study concept and design: HB and XY, Acquisition of data: HB and MNK, Analysis and interpretation of data: HB, MNK, and XY, Drafting of the manuscript: HB, MNK andTR., Critical revision of the manuscript for important intellectual content: XY, TR and MFH., Administrative, technical, material support, and institutional study supervision: XY. All the authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not Applicable.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Bilal, H., Khan, M.N., Rehman, T. et al. Antibiotic resistance in Pakistan: a systematic review of past decade. BMC Infect Dis 21, 244 (2021). https://doi.org/10.1186/s12879-021-05906-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-021-05906-1