Abstract
Alicyclobacillus acidoterrestris, an acidophilic and thermophilic bacteria, is an important microbial resource for stress resistance genes screening. In this study, DnaK gene from A. acidoterrestris was subcloned to construct the recombinant plasmid pET28a–DnaK. The successful construction of the plasmid was verified by double-enzyme digestion and sequencing analysis. The recombinant plasmid was transformed into Escherichia coli BL21 and isopropy-β-d-thiogalactoside (IPTG) was used to induce recombinant E. coli to express DnaK gene. A 70 kD fusion protein was identified by SDS-PAGE, which suggested that DnaK gene from A. acidoterrestris was successfully expressed. The recombinant and wild BL21 were treated with high temperatures of 54, 56 and 58 °C at pH values of 5.0–7.0 to compare the effects of heterogeneous expression of the DnaK gene from A. acidoterrestris on the stress resistance. The experimental results showed that survival rate of recombinant BL21–DnaK has been improved considerably under heat and acid stresses in contrast with the wild BL21, and D-values of recombinant BL21 were 14.7–72% higher than that of wild BL21, which demonstrated that heterogeneous expression of DnaK gene from A. acidoterrestris could significantly enhance the resistance of host bacteria E. coli against heat and acid stresses.
Similar content being viewed by others
Introduction
During fermentation production process, microbial cell could encounter stresses from various aggressive conditions including high temperature, increased ethanol concentrations, high acidity and osmotic pressures, which could inhibit cell viability, lead to cell death, result in fermentation abnormal ceasing and thus reduce production efficiency etc. (Shafiei 2013; Bubulya 2011; Logothetis et al. 2007). Therefore, a major focus in food microbial research is to investigate ways to improve stress resistance of microbes cell in various severe conditions and maintain the high productivity of microbial in the industrial environment (Sugimoto et al. 2010; Andersson et al. 2010).
Hsp70 possesses multiple biological functions and it has been well recognized as an induced chaperone protein by heat shock and other abiotic stresses, and DnaK can become over-expressed in cases of heat, high salt and alcohol to enhance tolerance of cell to harm and maintain normal metabolism (Groemping and Reinstein 2001; Seydlová et al. 2012; Di Pasqua et al. 2013). It has been shown that DnaK mRNA and DnaK protein of Streptococcus mutants were increased in response to acid shock (Jayaraman et al. 1997), and the strain’s capacity for growth in low-pH media becomes impaired, when DnaK levels of S. mutans are lowered via knockdown, strongly indicating the involvement of DnaK in acid tolerance (Lemos et al. 2007). Tomoyasu et al. (2012) found that DnaK knockout mutants of Streptococcus intermedius exhibited slow growth, thermosensitivity and accumulation of GroEL in the cell. The results imply that molecular chaperone most likely play a major role in the stress response of the bacteria.
Alicyclobacillus acidoterrestris is a genus of spoilage bacteria causing contamination in low pH foods such as apple juice and other beverage products due to its heat and acid resistance that enables survival from the traditional pasteurization procedures (90–95 °C for 30–60 s) (Smit et al. 2011). Due to the particular physiology, A. acidoterrestris has gained research interests focusing on detection and control in fruits juice (Mast et al. 2016; Cai et al. 2015; Bevilacqua et al. 2013), but gene regulation mechanism in the bacteria responding to high temperature and acidity are still unclear. It is reported that expression levels of DnaK gene in A. acidoterrestris were up-regulated rapidly under heat and acid stresses, and played significant roles for the survival of A. acidoterrestris in heat and acid conditions (Jiao et al. 2015). To validate the biological functions of DnaK gene in A. acidoterrestris resisting heat and acid stresses, recombinant Escherichia coli BL21 (DE3) expressing DnaK gene from A. acidoterrestris were constructed and the cell viability under heat and acid stresses were tested to evaluate effects of heterogeneous expression of the DnaK gene on stress resistance of recombinant E. coli BL21–DnaK, the objective of the study aims at the improvement of stress tolerance of microbes by transgenic technology in fermentation industry and provides theoretical basis for the development of the new microorganisms with high production and preferable stress tolerant properties.
Materials and methods
Bacterial strains and plasmids
The plasmid pET-28a(+) used for gene fusion construction was the pET System (Novagen, Germany). E. coli BL21 (DE3) strains were used as cloning hosts through this study. Alicyclobacillus acidoterrestris DSM 3922T was purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany).
Construction of recombinant E. coli BL21 expressing DnaK gene
Amplification of DnaK gene (GenBank Accession No. HQ893543) was performed by PCR using genomic DNA of A. acidoterrestris as the template with the 5′ end primer (dnaKbF: 5′-GAGCTCCCATGGAGGAGGAACTTTCAGTGGCAAAAG-3′) containing an initiator ATG incorporated into an NcoI-site (shown in italics) and with the 3′ end primer (dnaKbR:5′-GTCGACCTCGAGCTTCTGATCCTTGTCAACTTCGGTG-3′) to add an XhoI-site (in boldface type).
PCR amplification were conducted using the following protocol: 10 cycles of 94 °C for 5 min, 94 °C for 35 s, 61–55 °C for 30 s, 72 °C for 4 min, and 20 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 4 min and followed by 10 min at 72 °C. The PCR products were gel-purified and digested using NcoI/XhoI double digestion and subcloned into pET-28a(+) vector linearized by a double digestion with the same restriction enzymes to construct pET-28a(+)-DnaK recombinant plasmid, which was transformed into E. coli BL21. Bacterium liquid PCR and DNA sequencing were used to screen the positive recombinant clones.
Induction and expression of DnaK protein
Recombinant E. coli BL21 were cultured in Luria–Bertani (LB) broth containing 50 mg/mL of kanamycin monosulfate at 37 °C overnight. Fifteen microliters of the overnight cultures were added to 15 mL fresh LB containing 50 mg/mL of kanamycin monosulfate and continued to incubate for 2–3 h and shaken at frequency of 250 rpm at 37 °C until mid-log phase (OD600 = 0.6). Then, half of the cultures is put into fridge for storage at 4 °C and used as control group (non-induced sample). The other half culture (approximately 7 mL) is added with isopropyl β-d-thiogalactopyranoside (IPTG) solution to a final concentration of 1 mM and induced at 37 °C for 3 h. 5 mL induced E. coli BL21 cultures (were centrifuged at 3000×g) for 10 min to harvest cells and supernatant is used as sample for LB broth. Then, the cells were resuspended in 50 mM phosphate buffer (pH 8.0, 500 mM NaCl, 1 mM EDTA) and lysed by sonication and the cell lysate was separated by centrifugation at 3000×g for 10 min into soluble protein extracts and sediment used as sample for SDS-PAGE analysis.
Thermal inactivation of transformed E. coli at static temperatures
Cultures of E. coli BL21 were grown as described above. IPTG solution was added to mid-log phase cultures (OD600 = 0.6) to a final concentration of 1 mM, and incubation was continued at 37 °C for 3 h. After IPTG induction, the cells were centrifuged at 3000×g for 10 min to harvest the cells and then resuspended in fresh LB broth with different pH of 7.0, 6.0, 5.5 and 5.0, then immediately immersed in a water bath at 54, 56 and 58 °C respectively and treated for 10, 20, 30, 40 and 50 min.
The treated samples of 1 mL were taken every 10 min and decimal serial dilutions of the treated samples were prepared in a stroke-physiological saline solution and 100 μL of serial dilutions samples plated in triplicate onto LB plus kanamycin monosulfate plates. Plates were incubated for 24 h at 37 °C and colony forming units were enumerated to estimate cell viability (Velliou et al. 2011).
Data analysis
Each sample collected in the experiment was analyzed in three replicates. Means of data were analyzed and compared by T test at P < 0.01 using statistical software (SAS version 9.4, USA). The experimental data (bacteria colonies number) were log-transformed and plotted as a function of time. Equation 1 was obtained by linear fitting and D-value was calculated.
with N (t) [Log (CFU/mL)] the stress resistant subpopulation, N (0) [Log (CFU/mL)] the initial cell population, t [min] the time and D [min] the time required to kill 90% of the bacteria.
Results
Construction of recombinant E. coli strain expressing DnaK gene
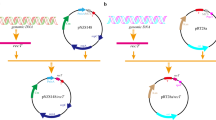
The coding region of DnaK gene was amplified with two primers designed and cloned into the pET-28a(+) plasmid to construct pET-28a(+)-DnaK expression vector for fusion protein expression. Then the recombinant plasmid pET-28a (+)-DnaK (Fig. 1) was transformed into E. coli and 3 clones were selected by growth in kanamycin monosulfate-containing medium.
The clones containing recombinant plasmid were identified by PCR, and then confirmed by double enzyme digestion with NcoI and XhoI and nucleotide sequencing. As shown in Fig. 1, the length of all PCR products was approximately 1854 bp and target fragment is inserted into vector with accurate enzyme digestion site (Fig. 2).
Based on nucleotide sequencing of the expression vectors produced, one clone (E. coli BL21–DnaK) was selected for expression of the recombinant protein. At the same time, the pET-28a(+) empty vector alone was also transformed into BL21 as the control group.
Induction of fusion protein in recombinant E. coli cells
The supernatants of sonicated cells lysates from the 1 mM IPTG induced transformed cells were investigated to detect the presence of fusion protein by SDS–PAGE analysis. According to the construction (Fig. 1), the transformed E. coli cells should produce 70 kD fusion protein. A band about 70 kD fusion protein was clearly observed in gels containing cell supernatant, indicating this vector works normally and DnaK protein was expressed in the transformed E. coli BL21cells after the 3 h induction of IPTG (Fig. 3).
SDS–PAGE analysis of DnaK protein expression in recombinant E. coli BL21–DnaK. Lane M low molecular weight protein standard; lane 1 whole-cell proteins of uninduced recombinant E. coli BL21–DnaK; lane 2 whole-cell proteins of induced recombinant E. coli BL21–DnaK; lane 3 supernatant of induced cells lysate; lane 4 precipitation of induced cells lysate; lane 5 LB medium
Moreover, content of soluble protein exceeds that of insoluble protein by a large margin and target protein is not detected in the supernatant of LB culture medium, which showed that DnaK fusion protein is mainly expressed as intracellular and soluble protein with no inclusion formation even accumulated at 37 °C.
Stress resistance of recombinant E. coli cells expressing DnaK gene
In order to evaluate effects of induced expression of DnaK gene on stress resistance of host bacteria, recombinant E. coli BL21 expressing DnaK gene together with wild E. coli BL21 is treated with heat and acid stresses. Cell viability was measured by counting colony-forming units in triplicate LB plates and D-values were calculated using Eq. 1 to compare the heat resistance of recombinant E. coli BL21–DnaK with that of wild E. coli BL21 (Fig. 4).
D-values of recombinant E. coli BL21–DnaK and E. coli BL21 at heat and acid stresses. a pH 5.0, b pH 5.5, c pH 6.0, d pH 7.0. The results represent the mean ± SD of three biological replicates. T-test is used to analyze the statistical differences. Different capital letters on the top of each column indicate the differences at the significant level of 0.01 between groups
As shown in Fig. 4, heat and acid stresses have significant effects on survive of wild and recombinant E. coli and result in declines of D-values, which implicated a quicker inactivation. The D-values for BL21–DnaK and BL21 in LB medium at different combination of temperature and pH-value ranged from 16.15 to 8.72 min and from 14.08 to 5.2 min, respectively. At the low pH of 5.0, D-values of E. coli BL21–DnaK were 25.69, 50.08 and 67.69% higher than that of wild E. coli BL21 under the heat stress of 54, 56, and 58 °C, respectively. Under the other treatment conditions of the same pH value and temperature, D-values of E. coli BL21–DnaK were higher than that of wild E. coli BL21 and ranged from 14.7 to 72%. The experiment suggested that DnaK gene expression not only led to a higher heat resistance of recombinant E. coli BL21–DnaK, but also significantly improved the resistance against acid stress.
Discussion
For fermentation industry, the improvement of strain resistance against high temperature and acid could considerably reduce cooling requirements and prevent contaminative microbes, which avoided consumption of water and other resources and enhanced productivity, having important application significance and considerable commercial value (Gupta and Mukerji 2001). However, in adverse conditions of high temperature and strong acidity, a great many proteins in the microbe could be degenerated or aggregated, and finally led to the death of industrial microorganisms (Tissiéres et al. 1974). Hence, a key factor to improve the survival capacity and resistance of organism in stress conditions such as heat and acid is to prevent denaturation of intracellular proteins. Heat shock proteins could identify hydrophobic area on the surface of denatured protein, accelerate new protein folding and prevent irreversible agglutination reaction, thus effectively protect cell before damage (Horwitz 1992).
Extreme microorganisms in various habitats respond to external severe conditions by adapting their physiology through gene expression in order to survive and contain many unique functioning proteins itself. Research results showed that over expression of recombinant prefoldin deriving from Pyrococcus horikoshii OT3 could give E. coli tolerance of some organic solvents (Okochi et al. 2008), which suggested that heterogeneous expression of recombinant molecular chaperone gene from Themophiles played critical roles in enhancing the ability of host bacteria to resist the hostile circumstance. The heat shock protein genes have been successfully separated and researches have been conducted on inducing factors for the expressions and the relationship between over expression of heat shock proteins and heat resistance of organisms (Yeh et al. 1997).
Hsp70s, generally called chaperone protein DnaK in bacteria, were first discovered in fruit flies in the 1960s (Ritossa 1962) and responsible for essential functions in response to various biotic and abiotic stresses and involved in many cellular processes, including protein folding, protein translocation across membranes and regulation of protein degradation, which has been a hot research area in recent years (Yu et al. 2015). Studies in Staphylococcus aureus indicated that the DnaK system contributes to heat and oxidative stress tolerance, and DnaK protein refolding machinery plays an essential role in the stress responses (Singh et al. 2007). According to Simon et al. (1995), over expression of HSP70 could improve survival rates of murine fibroblasts exposed in ultraviolet rays and protect cells against DNA damage caused by ultraviolet C (Niu et al. 2006). Laksanalamai et al. (2006) found that molecular chaperone of extremely thermophilic bacteria P. furiosus had significant protective action to polymerase of Taq DNA under high temperature, which indicated that expression of chaperone extremophiles could improve tolerance of host cell against severe external environment.
Interest in the Hsp70 class of chaperones is growing because of the large variety of cellular processes in which they are involved, as well as their possible participation in environmental stress in vitro and pathological and physiological stresses in vivo (Gong and Golic 2006). For the objective of investigating the effects of heterogeneous expression of DnaK gene from A. acidoterrestris on stress resistance of E. coli, the research has established the expression carrier pET28a–DnaK, transformed into E. coli BL21, and then cell viability and resistance of recombinant and wild E. coli BL21 to high temperature and acidity was analysed and expressed in decimal reduction time (D-value). The research results showed that DnaK gene after induction of IPTG demonstrate a significant improvement of resistance of recombinant E. coli BL21–DnaK against high temperature and acidity, which mainly indicated by considerable increases in surviving rate and D-values of recombinant BL21–DnaK comparing to wild BL21. In microbiology, D-value refers to decimal reduction time and is the time required at a given temperature to kill 90% of the exposed microorganisms (Mazzola et al. 2003). Therefore, D-value of microorganisms is usually used in assessing microbial thermal resistance and thermal death time analysis (Berendsen et al. 2015). In our study, under heat stress at the same temperature, the D-values of microorganism decreased with declining pH-values. Moreover, under acid stress at the same pH-value, the D-values decreased with increasing temperature, which signifies that the higher the temperature, the faster the inactivation. The results of our study were in accordance with the general rules about effects of temperature and pH value on the thermal properties of microorganisms. However, the D-values of recombinant BL21–DnaK were 14.7 to 72% higher than that of wild BL21 under the same treatment temperature and acidity, which indicated DnaK gene expression in host bacteria E. coli BL21 could lead to a higher heat and acid resistance.
The DnaK gene is constitutive expression in A. acidoterrestris at the normal growth temperature of 45 °C, its expression is rapidly increased by 40% at a heat shock temperature of 70 °C for 5 min and induced 4.3-fold by acid stress in the low pH of 1.0 for 1 h (Jiao et al. 2015). The prominent expression of Dnak gene suggests that it may play an important role in thermotolerance and acid resistivity of A. acidoterrestris, and the results in the further study confirmed this supposition, which showed that heterogeneous expression of the Dnak gene could improve heat and acid resistance of recombinant E. coli. Changes in gene expression constitute the main component of the bacterial response to stress and environmental changes and molecular chaperones proteins are one of the main mechanisms (De Bruijn 2016). The function of Dnak gene from A. acidoterrestris improving by a large margin resistance of the host bacterium against acid and heat stresses might be related with special physiological potency of A. acidoterrestris, but the regulating mechanism needs further research to fully elucidate the roles of Dnak gene, the results of this study could contribute to the development of high endurance fermentation strains against adverse environments such as high temperature and acid in food industry.
Abbreviations
- A. acidoterrestris :
-
Alicyclobacillus acidoterrestris
- E. coli :
-
Escherichia coli
- IPTG:
-
isopropy-β-d-Thiogalactoside
- SDS-PAGE:
-
SDS-polyacrylamide gelelectrophoresis
References
Andersson C, Petrova E, Berglund K, Rova U (2010) Maintaining high anaerobic succinic acid productivity by product removal. Bioproc Biosyst Eng 33:711–718. doi:10.1007/s00449-009-0393-y
Berendsen EM, Zwietering MH, Kuipers OP, Wells-Bennik MH (2015) Two distinct groups within the Bacillus subtilis group display significantly different spore heat resistance properties. Food Microbiol 45:18–25. doi:10.1016/j.fm.2014.04.009
Bevilacqua A, Campaniello D, Speranza B, Sinigaglia M, Corbo MR (2013) Control of Alicyclobacillus acidoterrestris in apple juice by citrus extracts and a mild heat-treatment. Food Control 31:553–559. doi:10.1016/j.foodcont.2012.12.014
Bubulya P (2011) Cell metabolism–cell homeostasis and stress response. In: Sousa MJ, Ludovico P, Rodrigues F, Leão C, Côrte-Real M (eds) Stress and cell death in yeast induced by acetic acid. InTech, Croatia, pp 73–100
Cai R, Wang ZL, Yuan YH, Liu B, Wang L, Yue TL (2015) Detection of Alicyclobacillus spp. in fruit juice by combination of immunomagnetic separation and a SYBR green I real-time PCR assay. PLoS ONE 11:e0146556. doi:10.1371/journal.pone.0146556
De Bruijn FJ (2016) Stress and environmental regulation of gene expression and adaptation in bacteria. Wiley, Hoboken
Di Pasqua R, Mauriello G, Mamone G, Ercolini D (2013) Expression of DnaK, HtpG, GroEL and Tf chaperones and the corresponding encoding genes during growth of Salmonella thompson in presence of thymol alone or in combination with salt and cold stress. Food Res Int 52:153–159. doi:10.1016/j.foodres.2013.02.050
Gong WJ, Golic KG (2006) Loss of Hsp70 in Drosophila is pleiotropic, with effects on thermotolerance, recovery from heat shock and neurodegeneration. Genetics 172:275–286. doi:10.1534/genetics.105.048793
Groemping Y, Reinstein J (2001) Folding properties of the nucleotide exchange factor GrpE from Thermus thermophilus: GrpE is a thermosensor that mediates heat shock response. J Mol Biol 314:167–178. doi:10.1006/jmbi.2001.5116
Gupta R, Mukerji KG (2001) Microbial technology. APH Publishing Corporation, New Delhi
Horwitz J (1992) Alpha-crystallin can function as a molecular chaperone. Proc Natl Acad Sci 89:10449–10453. doi:10.1073/pnas.89.21.10449
Jayaraman GC, Penders JE, Burne RA (1997) Transcriptional analysis of the Streptococcus mutans hrcA, grpE and dnaK genes and regulation of expression in response to heat shock and environmental acidification. Mol Microbiol 25:329–341. doi:10.1046/j.1365-2958.1997.4671835.x
Jiao LX, Ran JJ, Xu XX, Wang JJ (2015) Heat, acid and cold stresses enhance the expression of DnaK Gene in Alicyclobacillus acidoterrestris. Food Res Int 67:183–192. doi:10.1016/j.foodres.2014.11.023
Laksanalamai P, Pavlov AR, Slesarev AI, Robb FT (2006) Stabilization of Taq DNA polymerase at high temperature by protein folding pathways from a hyperthermophilic archaeon, Pyrococcus furiosus. Biotechnol Bioeng 93:1–5. doi:10.1002/bit.20781
Lemos JA, Luzardo Y, Burne RA (2007) Physiologic effects of forced down-regulation of dnaK and groEL expression in Streptococcus mutans. J Bacteriol 189:1582–1588. doi:10.1128/jb.01655-06
Logothetis S, Walker GM, Nerantzis ET (2007) Effect of salt hyperosmotic stress on yeast cell viability. Proc Natl Sci Matica Srpska Novi Sad 113:271–284. doi:10.2298/zmspn0713271l
Mast S, Dietrich R, Didier A, Märtlbauer E (2016) Development of a polyclonal antibody-based sandwich enzyme-linked immunosorbent assay for the detection of spores of Alicyclobacillus acidoterrestris in various fruit juices. J Agric Food Chem 64:497–504. doi:10.1021/acs.jafc.5b03841
Mazzola PG, Penna TCV, da S Martins AM (2003) Determination of decimal reduction time (D value) of chemical agents used in hospitals for disinfection purposes. BMC Infect Dis 3:1. doi:10.1186/1471-2334-3-24
Niu P, Liu L, Gong ZY, Tan H, Wang F, Yuan J, Feng YM, Wei QY, Tanguay RM, Wu TC (2006) Overexpressed heat shock protein 70 protects cells against DNA damage caused by ultraviolet C in a dose-dependent manner. Cell Stress Chaperon 11:162–169. doi:10.1379/csc-175r.1
Okochi M, Kanie K, Kurimoto M, Yohda M, Honda H (2008) Overexpression of prefoldin from the hyperthermophilic archaeum Pyrococcus horikoshii OT3 endowed Escherichia coli with organic solvent tolerance. Appl Microbiol Biotechnol 79:443–449. doi:10.1007/s00253-008-1450-1
Ritossa F (1962) A new puffing pattern induced by temperature shock and DNP in Drosophila. Experientia 18:571–573. doi:10.1007/bf02172188
Seydlová G, Halada P, Fišer R, Toman O, Ulrych A, Svobodová J (2012) DnaK and GroEL chaperones are recruited to the Bacillus subtilis membrane after short-term ethanol stress. J Appl Microbiol 112:765–774. doi:10.1111/j.1365-2672.2012.05238.x
Shafiei R (2013) Fundamental and applied studies on freeze-dried Vinegar Starter. Doctoral dissertation, Université de Liège
Simon MM, Reikerstorfer A, Schwarz A, Krone C, Luger TA, Jäättelä M, Schwarz T (1995) Heat shock protein 70 overexpression affects the response to ultraviolet light in murine fibroblasts. Evidence for increased cell viability and suppression of cytokine release. J Clin Invest 95:926–933. doi:10.1172/jci117800
Singh VK, Utaida S, Jackson LS, Jayaswal RK, Wilkinson BJ, Chamberlain NR (2007) Role for dnaK locus in tolerance of multiple stresses in Staphylococcus aureus. Microbiol 153:3162–3173. doi:10.1099/mic.0.2007/009506-0
Smit Y, Cameron M, Venter P, Witthuhn RC (2011) Alicyclobacillus spoilage and isolation—a review. Food Microbiol 28:331–349. doi:10.1016/j.fm.2010.11.008
Sugimoto S, Higashi C, Matsumoto S, Sonomoto K (2010) Improvement of multiple-stress tolerance and lactic acid production in Lactococcus lactis NZ9000 under conditions of thermal stress by heterologous expression of Escherichia coli dnaK. Appl Environ Microbiol 76:4277–4285. doi:10.1128/aem.02878-09
Tissiéres A, Mitchell HK, Tracy UM (1974) Protein synthesis in salivary glands of Drosophila melanogaster: relation to chromosome puffs. J Mol Biol 84:389–398. doi:10.1016/0022-2836(74)90447-1
Tomoyasu T, Tabata A, Imaki H, Tsuruno K, Miyazaki A, Sonomoto K, Nagamune H (2012) Role of Streptococcus intermedius DnaK chaperone system in stress tolerance and pathogenicity. Cell Stress Chaperon 17:41–55. doi:10.1007/s12192-011-0284-4
Velliou EG, Van Derlinden E, Cappuyns AM, Nikolaidou E, Geeraerd AH, Devlieghere F, Van Impe JF (2011) Towards the quantification of the effect of acid treatment on the heat tolerance of Escherichia coli K12 at lethal temperatures. Food Microbiol 28:702–711. doi:10.1016/j.fm.2010.06.007
Yeh CH, Chang PFL, Yeh KW, Lin WC, Chen YM, Lin CY (1997) Expression of a gene encoding a 16.9-kDa heat-shock protein, Oshsp16.9, in Escherichia coli enhances thermotolerance. Proc Natl Acad Sci 94:10967–10972. doi:10.1073/pnas.94.20.10967
Yu A, Li P, Tang T, Wang J, Chen Y, Liu L (2015) Roles of Hsp70s in stress responses of microorganisms, plants, and animals. Biomed Res Int 2015:1–8. doi:10.1155/2015/510319
Authors’ contributions
LJ was primarily responsible for the designing experimental program. The main work of author XX was construction of recombinant E. coli BL21 and expression of DnaK gene in E. coli. XF was responsible for detection of resistance of recombinant and wild E. coli BL21 to high temperature and acidity. Data analysis was accomplished by JR. XL and RZ offered some valuable thoughts and advice for modifying the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Funding
This study was funded by National Natural Science Foundation of China (31401673) and Program for Science and Technology Innovation Talents in Universities of Henan Province (17HASTIT037).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Xu, X., Jiao, L., Feng, X. et al. Heterogeneous expression of DnaK gene from Alicyclobacillus acidoterrestris improves the resistance of Escherichia coli against heat and acid stress. AMB Expr 7, 36 (2017). https://doi.org/10.1186/s13568-017-0337-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13568-017-0337-x