Abstract
Background
Blood parasites of the genus Haemoproteus Kruse, 1890 are cosmopolitan, might be responsible for mortality in non-adapted birds, and often kill blood-sucking insects. However, this group remains insufficiently investigated in the wild. This is particularly true for the parasites of leaf warblers of the Phylloscopidae Alström, Ericson, Olsson & Sundberg the common small Old World passerine birds whose haemoproteid parasite diversity and vectors remain poorly studied. This study reports a new species of Haemoproteus parasitizing leaf warblers, its susceptible vector and peculiar phylogenetic relationships with other haemoproteids.
Methods
Wood warblers (Phylloscopus sibilatrix Bechstein) were caught in Lithuania during spring migration, and blood films were examined microscopically. Laboratory reared Culicoides nubeculosus Meigen were exposed experimentally by allowing them to take blood meals on one individual harbouring mature gametocytes of the new Haemoproteus species (lineage hPHSIB2). To follow sporogonic development, the engorged insects were dissected at intervals. The parasite lineage was distinguished using sequence data, and morphological analysis of blood and sporogonic stages was carried out. Bayesian phylogeny was constructed in order to determine the phylogenetic relationships of the new parasite with other haemoproteids.
Results
Haemoproteus (Parahaemoproteus) homopalloris n. sp. was common in wood warblers sampled after arrival to Europe from their wintering grounds in Africa. The new parasite belongs to a group of avian haemoproteid species with macrogametocytes possessing pale staining cytoplasm. All species of this group clustered together in the phylogenetic analysis, indicating that intensity of the cytoplasm staining is a valuable phylogenetic character. Laboratory-reared biting midges C. nubeculosus readily supported sporogony of new infections. Phylogenetic analysis corroborated vector experiments, placing the new parasite in the clade of Haemoproteus (Parahaemoproteus) parasites transmitted by biting midges.
Conclusions
Haemoproteus homopalloris n. sp. is the third haemoproteid, which is described from and is prevalent in wood warblers. Phylogenetic analysis identified a clade containing seven haemoproteids, which are characterised by pale staining of the macrogametocyte cytoplasm and with ookinetes maturing exceptionally rapidly (between 1 to 1.5 h after exposure to air). Both these features may represent valuable phylogenetic characters. Studies targeting mechanisms of sporogonic development of haemoproteids remain uncommon and should be encouraged. Culicoides nubeculosus is an excellent experimental vector of the new parasite species.
Similar content being viewed by others
Background
Blood parasites of the genus Haemoproteus Kruse, 1890 (Haemosporida: Haemoproteidae) are distributed worldwide and are among the most extensively studied blood parasites of birds, particularly in the temperate regions [1]. They are transmitted by biting midges (Ceratopogonidae) and louse flies (Hippoboscidae) [2]. Haemoproteus species were considered relatively benign to their avian hosts [3]. However, several recent studies demonstrated the negative influence of these parasites and even mortality due to haemoproteosis not only in non-adaptative birds [4,5,6,7,8,9,10,11], but also in blood-sucking insects [12, 13].
Numerous studies addressed morphological and molecular characterization, distribution and genetic diversity of haemoproteids [14,15,16,17,18,19,20]. However, some bird groups remain insufficiently investigated. This is particularly true for leaf warblers of genus Phylloscopus Boie belonging to the Phylloscopidae Alström, Ericson, Olsson & Sundberg. The wood warbler, Phylloscopus sibilatrix Bechstein, is a common European passerine bird wintering in sub-Saharan Africa [21]. Despite its broad range of occurrence, only two haemoproteid species, Haemoproteus majoris Laveran, 1902 [2, 22,23,24] and Haemoproteus belopolskyi Valkiūnas, 1989 [25], have been reported in this bird.
Several research groups successfully used molecular techniques to detect haemoproteids in wild caught insects [26,27,28,29,30,31]. PCR-based protocols detect the presence of parasite DNA in the insect, but do not provide information on which development stage the parasite is present. In other words, these tools are insufficiently sensitive to conclude if invasive stages (sporozoites) develop in the PCR-positive insect and if insects can act as vectors and transmit the parasites [32]. Observation of infective sporozoites in salivary glands strongly suggests the vectorial capacity of blood-sucking insects. Experimental infections provide the opportunity to follow parasite development and to morphologically characterize each life stage [33, 34].
Studies addressing the sporogonic development and transmission of avian Haemoproteus species are few [29, 35,36,37,38]. Vector species and complete life-cycle remains unknown for the great majority of Haemoproteus parasites and their lineages [1, 38, 39]. Culicoides species have been used in experimental studies addressing parasite development [33, 34]. These insects are abundant in mixed forest zone in eastern Europe [37]. Despite of their diminutive size and difficulties to maintain in colonies [40], Culicoides nubeculosus Meigen and several other species have been kept in laboratory colonies successfully [41, 42]. Culicoides nubeculosus has been used to study the sporogonic development of Haemoproteus handai Maqsood, 1943 [43], Haemoproteus tartakovskyi Valkiūnas, 1986 [34], Haemoproteus noctuae Celli & Sanfelice, 1891 and Haemoproteus syrnii Mayer, 1910 [33].
During this study, we discovered a new Haemoproteus species that infects wood warblers. This parasite is described using morphology of blood stages and molecular data of partial cytochrome b (cytb) gene sequence. To access information about sporogonic development, we experimentally infected the biting midge Culicoides nubeculosus. The main objectives of this study were (i) to characterize the new Haemoproteus species morphologically; (ii) to develop its molecular characterization based on partial cytb sequence; (iii) to determine phylogenetically closely related parasite species; and (iv) to follow sporogonic development in experimentally infected vector.
Methods
Collection and examination of bird blood samples
We collected blood samples from 16 adult wood warblers (Phylloscopus sibilatrix) at the Ornithological Station in Ventės Ragas, Lithuania (55°20'28.1"N, 21°11'25.3"E) during May in 2015, 2016 and 2017. The birds were caught with mist nets. Approximately 30 μl of blood was withdrawn from the brachial vein using a sterile syringe needle and capillary tubes. Several drops were used immediately for preparation of blood smears on three glass slides, and the remaining blood was stored in SET buffer (0.05 M tris, 0.15 M NaCl, 0.5 M EDTA, pH 8.0) for molecular diagnostics. Blood smears were air-dried, fixed with absolute methanol, and stained with Giemsa [44]. Preparations of good quality and sufficient parasite intensity with single infection, as determined both by microscopic examination and PCR-based testing, were used for morphological characterization of the new species.
Olympus BX41 microscope equipped with PixeLINK and imaging software Megapixel FireWire Camera Release 3.2 were used to examine the blood films and prepare illustrations. Measurements were taken from the images using the calibrated Motic Images Plus 2.0. The slides were examined for 15–20 min at low magnification (×400), and then at least 100 fields were studied at high magnification (×1000). Parasite identification follows the guidelines of Valkiūnas [2]. All measurements are given in micrometres. Images of positive preparations were collected for measurement. Representative preparations were deposited in the Nature Research Centre, Vilnius, Lithuania (accession number 49021 NS and 49022 NS). The analyses were carried out using the Statistica 7 package. A parahapantotype blood film with gametocytes of closely related haemoproteid Haemoproteus palloris Dimitrov, Iezhova, Zehtindjiev, Bobeva, Ilieva, Kirilova, Bedev, Sjohölm & Valkiūnas, 2016 (deposited at Nature Research Centre, Vilnius, accession number 48832 NS) was used for comparisons with the new Haemoproteus species.
Parasitemia was estimated as a percentage by actual counting the number of parasites per 1000 red blood cells or per 10,000 red blood cells during light infections (i.e. < 0.1%) [45]. Morphology of gametocytes of the new species was also compared with the type-specimens deposited in Institute of Nature Research Centre, Vilnius, Lithuania, of Haemoproteus majoris (accession number 48893 NS, from Phylloscopus trochilus Linnaeus) and Haemoproteus belopolskyi (accession number 435.85p, from Hippolais icterina Vieillot) the only two Haemoproteus parasites reported in P. sibillatrix so far. Hapantotype material of the new species was also compared with other Haemoproteus species with pale staining macrogametocytes: Haemoproteus pallidus Valkiūnas & Iezhova, 1991 (accession number 963.89, from Ficedula hypoleuca Pallas), Haemoproteus pallidulus Križanauskienė, Pérez-Tris, Palinaukas, Hellgren, Bensch & Valkiūnas, 2010 (accession number 5420 NS, from Sylvia atricapilla Linnaeus), Haemoproteus minutus Valkiūnas & Iezhova, 1992 (accession number 245.85p, from Turdus merula Linnaeus), Haemoproteus concavocentralis Dimitrov, Zehtindjiev, Bensch, Ilieva, Iezhova & Valkiūnas, 2014 (accession number 48756 NS, from Coccothraustes coccothraustes Linnaeus) and Haemoproteus vacuolatus Valkiūnas, Iezhova, Loiseau, Chasar, Smith & Sehgal, 2008 (accession number 42415 NS, from Andropadus latirostris Strickland).
Sporogonic development experimental design
A naturally infected wood warbler with single infection with the new Haemoproteus lineage was used as a donor of gametocytes to expose Culicoides nubeculosus biting midges. The presence of a single infection in the donor bird was confirmed by microscopic examination (see above) and PCR-based testing, as described below. Insects were reared in the laboratory according to Boorman et al. [41]. Experimental procedures were according to Bukauskaitė et al. [13]. Briefly, biting midges were kept in card boxes covered with fine mesh bolting silk. For the experiment, a box with unfed insects was gently pressed to the feather-free area on pectoral muscles of infected bird. Culicoides nubeculosus took blood meals through the bolting silk, with the great majority of females being fully engorged within 30–40 min. Then, biting midges were transferred to a bigger cage made of bolting silk (12 × 12 × 12 cm), males and non-fed females were removed. Remaining insects were kept in a room with controlled temperature (22° C), relative humidity (75 ± 5%) and light-dark photoperiod of 17:7 h. Insects were supplied with a 10% sugar solution offered in cotton pads.
Dissection of biting midges, preparations of parasites and microscopic examination
Experimentally exposed biting midges were dissected, and preparation of ookinetes, oocysts and sporozoites were made. First, the insects were anesthetized by placing them in a tube covered with cotton-wool pads moistened with 96% ethanol. Biting midges were dissected on intervals in order to follow the development of the parasite in the insect. We examined midgut contents for ookinetes 0.5–12 h post exposure, midgut wall for oocysts 2–5 days post exposure (dpe), and salivary glands for sporozoites 7–8 dpe.
For visualizing ookinetes, midgut was dissected and gently crushed on the slide; the preparations were fixed and stained the same way as blood films. For oocyst observation, temporary preparations were made. Midguts were isolated on a glass slide and a drop of 2% mercurochrome solution was placed on the guts, which was then covered with coverslips. This simplified observation of oocysts. To visualize sporozoites, preparation was made by extracting the salivary glands from biting midges and gently crashing them to prepare small thin smears, which were fixed with absolute methanol and stained with 4% Giemsa solution for 1 h. After each insect dissection, residual parts of their bodies were fixed in 96% ethanol and used for PCR-based analysis to confirm presence of corresponding parasite lineage in vectors. Dissected needles were disinfected in fire to prevent contamination after each dissection.
All vector preparations were examined using Olympus BX43 light microscope equipped with Olympus SZX2-FOF digital camera and imaging software QCapture Pro 6.0, Image Pro Plus (Tokyo, Japan). All preparations were examined as described above for blood smears, and the representative preparations were deposited in the Nature Research Centre, Vilnius, Lithuania (accession numbers 49023 NS and 49024 NS).
DNA extraction, PCR, sequencing and phylogenetic analysis
Blood samples from the donor bird and residual parts of infected biting midges were examined for haemosporidian parasites by PCR amplification. Total DNA was extracted from both materials using a standard ammonium acetate method [46] and quantified by NanoPhotometer® P330 (IMPLEN). A nested PCR protocol was used to amplify a cytb gene fragment [47, 48]. The first pair of primers (HaemFNI/HaemNR3) amplifies sequences of Plasmodium, Haemoproteus and Leucocytozoon. The second pair of primers (HaemF/HaemR2) is specific for Plasmodium and Haemoproteus parasites. We performed PCR amplification in 25 μl total volume including 50 ng of total genomic DNA template (2 μl), 12.5 μl of Phusion High-Fidelity PCR Master Mix (Thermo Fisher Scientific, Vilnius, Lithuania), 8.5 μl nuclease-free water and 1 μl of each primer (10 μM concentration). One positive (infection confirmed by microscopy analysis) and one negative control (ultrapure water) were used. Positive results were visualized by electrophoresing 2 μl of the final PCR product on a 2% agarose gel. Amplicons of proper length (approximately 500 bp) were precipitated and sequenced from both ends using Big Dye Terminator V3.1 cycle Sequencing Kit and ABI PRISMTM 3100 capillary sequencing robot (Applied Biosystems, Foster City, California). Sequences were edited and aligned using the BioEdit program [49] and deposited in the GenBank database (accession number MH513601). The presence of double peaks in sequence chromatograms was considered a co-infection.
A Bayesian phylogeny of parasite lineages was constructed based on alignment of 45 cytb lineages (33 Haemoproteus spp. and 11 Plasmodium spp.) using MrBayes version 3.2 [50]. One lineage of Leucocytozoon sp. (lineage lSISKIN2) was used as the outgroup. All lineages were carefully selected based on studies that provided morphological identification of parasites. We used the General Time Reversible model (GTR) selected by the software jModelTest 2 [51] as the best-fit model under the Bayesian Information Criterion. Gaps and missing data in the alignment were discarded prior to analyses. Two simultaneous runs were conducted with a sample frequency of every 100th generation over 3 million generations. We discarded 25% of the trees as ‘burn in’ period. The remaining trees were used to construct a majority rule consensus tree. The phylogenies were visualized using Fig Tree 1.4 [52]. The codes of cytb lineages are given according to MalAvi database, with a letter ‘h’ starting codes of Haemoproteus spp. lineages and a letter ‘p’ starting codes of Plasmodium spp. lineages. The sequence divergence between different lineages was calculated using Jukes-Cantor model of substitution, with all substitutions weighted equally (uniform rates), implemented in the program MEGA7 [53].
Results
Microscopic and molecular analysis of blood samples
In all, 43.8% of wood warbler tested by PCR and microscopy were infected with Haemoproteus parasites. All infections were detected by both methods equally. We reported two different lineages of Haemoproteus species in tested birds: hPHSIB1 (Haemoproteus majoris) and hPHSIB2 (Haemoproteus homopalloris n. sp.). The prevalence of infection with lineage hPHSIB2 was 31%. Two individuals harboured double haemosporidian infection, one with two Haemoproteus species, and another one with Haemoproteus and Plasmodium species. These double infections were detected also by PCR.
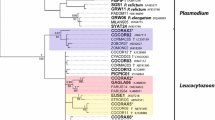
Phylogenetic analysis confirmed that morphological characteristics of gametocytes are of phylogenetic value because the lineage hPHSIB2 was readily distinguishable both morphologically (Fig. 1) and in the tree (Fig. 2). The lineage hPHSIB2 clustered with other so-called pale staining haemoproteid species, with a high (100%) posterior probability (Fig. 2, clade B): Haemoproteus pallidus, H. palloris, H. minutus, H. pallidulus, H. vacuolatus and Haemoproteus concavocentralis. It is worth noting that the mean genetic distance among cytb lineages within this clade was low (1.5%), but all parasites in this clade have unique morphological characters, based on which they can be distinguished from each other (see Fig. 3 and the Remarks below).
Gametocytes of two species of haemoproteids described from leaf warbles, Phylloscopidae. Haemoproteus homopalloris n. sp. (a-l) and Haemoproteus palloris (m-p). Young gametocytes (a, b), macrogametocytes (c-g, m, n) and microgametocytes (h-l, o, p). Long arrows: gametocyte nuclei; short arrows: vacuole-like spaces in macrogametocytes; arrowheads: pigment granules. Giemsa-stained thin blood films. Scale-bar: a-p, 10 μm
Bayesian phylogenetic inference of cytb gene lineages (479 bp) of 35 Haemoproteus spp. The tree is rooted with Leucocytozoon sp. (lineage lSISKIN2). Clades A and B indicate species of the subgenus Parahaemoproteus (a) and haemoproteids with pale-staining cytoplasm of gametocytes (b). MalAvi lineage codes are provided, followed by parasite species names and GenBank accession numbers. Nodal support values indicate Bayesian posterior probabilities. New species is given in bold
Haemoproteus spp. with pale staining of macrogametocyte cytoplasm. Haemoproteus concavocentralis (a-d), H. minutus (e-h), H. pallidus (i-l), H. pallidulus (m-p) and H. vacuolatus (q-t). Macrogametocytes (a, b, e, f, i, j, m, n, q, r), microgametocytes (c, d, g, h, k, l, o, p, s, t). Note the following valuable diagnostic features of the parasites: presence of a space between the nucleus of the infected erythrocyte and the growing gametocyte in H. concavocentralis (a); clearly irregular outline of mature gametocytes, which do not touch the poles of infected erythrocytes in H. minutus (e-h); gametocyte which are closely appressed to the nucleus of erythrocyte but do not touch the envelope of erythrocyte along their entire margin in H. pallidus (j, l); small pigment granules in mature gametocytes of H. pallidulus (m-p); presence of one prominent vacuole in the cytoplasm of each advanced macrogametocyte in H. vacuolatus (q-t). All these features are not characteristics of H. homopalloris n. sp. (see Fig. 1). Long simple arrows: gametocyte nuclei; short simple arrows: vacuole-like spaces in macrogametocytes; arrowheads: pigment granules; long simple wide arrows: space present between the parasite and an infected erythrocyte nucleus (a, d) and space between the parasite and the envelope of infected erythrocyte (j, l). Giemsa-stained thin blood films. Scale-bar: a-t, 10 μm
ᅟ
Family Haemoproteidae Doflein, 1916
Genus Haemoproteus Kruse, 1890
ᅟ
Haemoproteus ( Parahaemoproteus ) homopalloris n. sp.
ᅟ
Type-host: Phylloscopus sibilatrix Bechstein, 1793 (Passeriformes, Phylloscopidae), wood warbler.
Type-locality: Ornithological Station in Ventės Ragas (55°20'28.1"N, 21°11'25.3"E), Lithuania.
Type-specimens: Hapantotypes (accession numbers 49021 NS and 49022 NS, adult bird Phylloscopus sibilatrix; parasitaemia 0.1%, 5.vi.2017, Ornithological Station Ventės Ragas, collected by M. Ilgūnas) were deposited in the Institute of Ecology of Nature Research Centre, Vilnius, Lithuania. Parahapantotype (accession number G466204, other data as for the hapantotype) was deposited in the Queensland Museum, Brisbane, Australia. Co-infection with microfilaria is present in the type-material.
Site of infection: Mature erythrocytes; no other data.
Prevalence: 31% (5 out of 16 examined wood warblers were infected).
Representative DNA sequence: Mitochondrial cytb lineage hPHSIB2 (478 bp, GenBank accession number MH513601).
Vector: Sporogony completed and sporozoites developed in experimentally infected biting midges Culicoides nubeculosus. This insect is a convenient experimental vector. Natural vectors remain unknown. Representative preparations of sporogonic stages are deposited in the Institute of Ecology of Nature Research Centre, Vilnius, Lithuania, with the accession numbers of 49023 NS and 49024 NS.
ZooBank registration: To comply with the regulations set out in article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN) [54], details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) of the article is urn:lsid:zoobank.org:pub:AC9794B3-D735-4D36-BB6E-5CDF1CF2BA3F. The LSID for the new name Haemoproteus (Parahaemoproteus) homopalloris is urn:lsid:zoobank.org:act:5481116F-3F96-40D2-956E-96C710C64F29.
Etymology: The species name refers to the morphological and morphometric similarity of the new species with Haemoproteus palloris, a closely related haemoproteid infecting a closely related avian host, the willow warbler Phylloscopus trochilus.
Description (Fig. 1a-l, Table 1)
Young gametocytes. Rarely seen in the type-material (Fig. 1a, b). Elongate, with even outline and prominent pigment granules. Develop in mature erythrocytes; advanced growing gametocytes closely adhere to erythrocyte nuclei and extend longitudinally along nuclei.
ᅟ
Macrogametocytes. Develop in mature erythrocytes. Cytoplasm staining pale-blue, heterogeneous in appearance, lacking volutin granules. Outline even or slightly wavy (Fig. 1c-g). Vacuoles or vacuole-like spaces absent in the cytoplasm. Gametocytes grow along nuclei of infected erythrocytes, enclose nuclei with their ends, but do not encircle them completely (Fig. 1c-g). Advanced and fully grown macrogametocytes closely appressed both to nuclei and envelop of host cell. Fully grown gametocytes fill erythrocytes up to their poles, not displacing or only slightly displacing nuclei of infected cells laterally. Parasite nucleus relatively small (Table 1), of variable form and position; usually in subterminal position in gametocytes (Fig. 1c, d, f), but also observed in strictly terminal position (Fig. 1e, g) in 12% of macrogametocytes, a characteristic feature of this species development. Nucleolus not seen. Pigment granules roundish or oval, predominantly of medium size (0.5–1.0 μm), usually randomly scattered throughout the cytoplasm. Influence of gametocytes on host cell is non-pronounced (Table 1).
ᅟ
Microgametocytes. General configuration as in macrogametocytes with the usual haemosporidian sexual dimorphic characters, i.e. with large diffuse nuclei and relatively pale staining of the cytoplasm (Fig. 1h-l). Outline often even, but markedly irregular terminal gametocyte edges were also commonly observed (Fig. 1h).
Remarks
So far, H. homopalloris has only been recorded in Phylloscopus sibilatrix. One sequence with 100% similarity is deposited in GenBank (accession KJ488925), it was also reported in P. sibilatrix, in Western Greater Caucasus [55].
A characteristic feature of H. homopalloris n. sp. is the relatively pale staining of the cytoplasm in macrogametocytes, so that macro- and microgametocytes are relatively poorly distinguishable based on this character (compare Fig. 1f and Fig. 1i or l). Gametocytes with pale staining cytoplasm have been described in several species of Haemoproteus [2, 56,57,58]. These parasites seem to be common in African birds and can be often encountered in migrant European birds wintering in Africa [2, 56,57,58]. The pale staining cytoplasm is particularly different in cases of co-infections with two species (one with pale-stained and second with dark-stained macrogametocytes) present in same blood films.
Among the haemoproteids of passerine birds, H. homopalloris n. sp. is most similar to Haemoproteus palloris, lineage hWW1 (Fig. 1m-p). These parasites develop in closely related avian hosts, the wood warbler and the willow warbler Phylloscopus trochilus, so should be distinguished. They can be differentiated due to the presence of vacuoles or vacuole-like spaces in the majority (80%) of the advanced macrogametocytes of H. palloris (Fig. 1m, n) whereas such structures are absent in gametocytes of H. homopalloris n. sp. The second taxonomically distinctive character is the pattern of growth of gametocytes in these two parasites. In H. palloris (Fig. 1o), an unfilled space is usually present between the gametocyte and the erythrocyte nucleus [15]. This is not characteristic for H. homopalloris n. sp. The presence of nuclei in strictly terminal position in 12% of H. homopalloris magrogametocytes is another difference between these two species since this has not been reported in H. palloris. Despite of the presence of distinguishing morphological features, gametocytes of H. palloris and H. homopalloris n. sp. are similar (compare Fig. 1c-l with Fig. 1m-p) and the genetic difference among these two lineages is small (1.1% or 5 bp in 479 bp of the cytb gene sequence).
Haemoproteus majoris (lineage hPHSIB1) has been often reported in wood warblers [22,23,24]. Phylogenetic analysis showed that cytb lineage hPHSIB1 is significantly divergent from the lineage hPHSIB2 of H. homopalloris n. sp. (4.5% difference in 17 bp of cytb gene fragment) and parasites are morphologically different (compare Fig. 1c-l with Fig. 4a-d). The most distinctive difference between H. majoris and H. homopalloris is the presence of dumbbell-shaped growing gametocytes in the former ([2], see Fig. 4a), but such gametocytes are absent in H. homopalloris n. sp. Additionally, fully grown gametocytes of H. majoris markedly displace host cell nuclei laterally (NDR = 0.4 ± 0.1) [2], but this is not a case for H. homopalloris n. sp. (NDR = 0.8 ± 0.1) (compare Fig. 1c-l with Fig. 4b-d).
Gametocytes of two species of haemoproteids, which have been reported in the wood warbler Phylloscopus sibilatrix. Macrogametocytes (a-c, e-g) and microgametocytes (d, h) of Haemoproteus majoris (a-d) and H. belopolskyi (e-h). Note that the intensity of staining of the cytoplasm is different in macro- and microgametocytes. Long arrows: gametocyte nuclei; short arrows: vacuole-like spaces in macrogametocytes; arrowheads: pigment granules. Giemsa-stained thin blood films. Scale-bar: a-h, 10 μm
Haemoproteus homopalloris n. sp. (hPHSIB2) can be also readily distinguished from other Haemoproteus species with pale staining macrogametocytes. Haemoproteus concavocentralis (hHAWF2) has a characteristic space between nucleus of infected erythrocyte and parasite in growing gametocytes [58] (see Fig. 3a-d). Haemoproteus minutus (hTURDUS2) possesses a clearly irregular outline, and it does not touch the poles of infected erythrocytes [2, Fig. 3e-h]. Haemoproteus pallidus (hPFC1) gametocytes are closely appressed to the nucleus of erythrocyte but usually do not touch the envelope of erythrocyte along their entire margin [2, Fig. 3i-l]. Gametocytes of Haemoproteus pallidulus (hSYAT3) possess small pigment granules even in mature gametocytes ([57], Fig. 3m-p). Haemoproteus vacuolatus (hANLAT2) possesses one prominent vacuole in the cytoplasm of each advanced macrogametocyte ([56], Fig. 3q, r). None of these features are characters of H. homopalloris n. sp.
Sporogony in biting midges
The presence of numerous sporozoites in salivary glands confirms that H. homopalloris n. sp. can complete sporogony in C. nubeculosus. Ookinetes were not reported in preparations, but zygotes were numerous 8 h post-exposure (Fig. 5a). Oocysts were seen in midgut in temporary preparations 4 dpe. Sporozoites were detected in salivary glands preparations 7 dpe (Fig. 5b). The sporozoites possess fusiform bodies with centrally located nuclei and approximately equally pointed ends (Fig. 5b, Table 1). The PCR-based analysis and sequencing confirmed the presence of hPHSIB2 lineage in experimentally infected biting midges at sporozoite stage.
Discussion
Haemoproteus homopalloris n. sp. is the third species of haemoproteid parasite identified in wood warblers. The infection prevalence was high in the studied bird population. It is interesting to note that all birds were caught with stationary mist nets. In other words, the birds flew in the nets themselves, so they were actively moving individuals and looked healthy from this point of view.
The new species clustered with seven Haemoproteus species, which all possess the pale staining cytoplasm in macrogametocytes in the phylogenetic analysis based on cytb data (compare Figs. 1 and 3 with Fig. 4). This is not characteristic of the great majority of haemosporidian parasites, in which the cytoplasm of macrogametocytes stains more intensively than in microgametocytes (Fig. 4) [59].
The pale staining of the cytoplasm in H. homopalloris n. sp. (Fig. 1) and related species (Fig. 3) might be due to the approximately similar density of organelles in the cytoplasm of macrogametocytes in comparison to microgametocytes, resulting in accumulation of similar amount of stain in both types of cells during staining procedures. Additional electron microscope studies are needed to test this hypothesis. Different cellular structure of macro- and microgametocytes can be explained from the point of view of life span of these cells. Numerous cell organelles are needed for a long survival of macrogametocyte cells, which are relatively long-lived cells in comparison to microgametocytes. After fertilization, each macrogametocyte, transforms to zygote and then the same cell develops into ookinete and further, to relatively long living oocyst [2]. Microgametocytes are relatively short-lived cells; they only produce microgametocytes, which develop within several minutes in vectors. This suggests that haemoproteids with pale staining of the macrogametocyte cytoplasm should develop fast during sporogony in comparison to the species with intense staining of the cytoplasm. Available experimental observations support this and show that complete ookinete formation of H. minutus and H. pallidus, which macrogametocytes are pale-stained (Fig. 1c-l and Fig. 3), occur within 1 to 1.5 h both in vitro and in vivo at 18 to 20° C [2, 38]. These parasites also have tiny ookinetes (approximately 10 μm in length), which probably facilitates their rapid movement. The biological meaning of this phenomenon remains unclear. Because pale staining parasites cluster together in the phylogenetic tree (Fig. 2), it is possible that the rapid development of ookinetes might also occur in all other species in this clade. There is no explanation why this pattern is so applicable for the group of pale staining haemoproteid parasites. However, this pattern does not apply for other species like H. majoris, Haemoproteus tartakovskyi and H. belopolskyi, in which macrogametocytes are densely stained with Giemsa (Fig. 4) and mature ookinetes develop relatively slowly (between 6–24 h at the same conditions) [2, 38].
Morphological descriptions of Haemoproteus homopalloris n. sp. species was accompanied by DNA sequence information and phylogenetic hypothesis for its relationship with already described parasites. It has been suggested [60] and then confirmed by several other studies [15, 58, 61] that Haemoproteus lineages, which differ over 5% in partial cytb sequence (479 bp), are likely to be morphologically differentiated and can be distinguished on gametocyte stage. However, it was also shown that some cytb lineages of Haemoproteus spp. with difference of just two nucleotides (or 0.7%) could be identified based on morphological features of their gametocytes ([56, 62]; this study). This is particularly recognisable in closely related species (Figs. 1 and 3) with pale staining of macrogametocyte cytoplasm [15, 56,57,58]. The greatest divergence among the seven described species of this group is 3.1%.
The lineage hPHSIB2 of H. homopalloris n. sp. clustered with other lineages of the subgenus Parahaemoproteus (Fig. 2, clade A) and with other pale staining parasites (Fig. 2, clade B). Interestingly, this group of parasites forms a congruent monophyletic group with low cytb genetic divergence (up to 3.1%), but with clearly distinguishable morphological features (Fig. 3). The majority of haemoproteid species with pale staining of macrogametocytes have been reported in European birds wintering in sub-Saharan Africa or resident bird species in this region [2, 15, 56,57,58]. It might be that they originally evolved in countries with warm climates where they are diverse. Interestingly, H. homopalloris n. sp. and H. minutus readily complete sporogony in European biting midges at relatively low temperatures (close to 20° C) ([38]; this study), an indication that there are no obstacles to their transmission in the Palaearctic from the point of view of vector availability and temperature needed for sporogony.
It is important to note that one species of haemoproteid with pale staining cytoplasm in macrogametocytes, H. minutus, causes mortality of captive parrots in Europe [9, 63]. The mortality is due to heart damage by tissue exo-erythrocytic meronts of this parasite, which abort development before development of parasitemia [11]. Haemoproteus minutus is broadly distributed in the Palaearctic region, and several closely related lineages of this pathogen have been identified [63]. It is possible that other closely related to H. minutus parasites of clade B (Fig. 2) also might be virulent in non-adapted avian hosts. It is not clear if H. homopalloris n. sp. is virulent as the former species; the pathology of this parasite should be investigated in the future.
In this study, two birds were co-infected with different haemosporidian species, and the double infections were detected by both PCR diagnostics and microscopic examination. Various combinations of mixed infections are common in wild naturally infected birds [58, 64,65,66,67,68]. It is important to note that presence of mixed infections often markedly vary among different bird populations. Such infections might predominate and represent more than 80% in some bird species [64]. Mixed infections have been reported to be highly virulent in avian hosts [68, 69]. However, the currently used general primers often underestimate the prevalence of mixed infections [65, 70, 71], and this calls for application of new PCR protocols, which are sensitive to detect mixed infections of Haemoproteus and Plasmodium parasites [72]. However, all currently available PCR-based protocols remain insensitive to read mixed infections of several haemosporidian species belonging to the same genus or subgenus, suggesting essential need of application of microscopic examination and morphological identification in parasite biodiversity studies in the wild [44].
Four Haemoproteus species can complete sporogony in C. nubeculosus. These are Haemoproteus handai, Haemoproteus noctuae, Haemoproteus syrnii and Haemoproteus tartakovskyi [33, 34, 43]. This study adds H. homopalloris n. sp. to this list of haemoproteid species transmitted by this biting midge and reinforces the importance of C. nubeculosus in experimental vector research. The pattern of sporogony in Culicoides impuctatus seems to be similar in different Haemoproteus species under experimental conditions. During sporogonic development of H. balmorali Peirce, 1984, H. belopolskyi, H. dolniki Valkiūnas & Iezhova, 1992, H. fringillae Labbé, 1894, H. lanii Mello, 1936, H. minutus, H. parabelopolskyi Valkiūnas, Križanauskienė, Iezhova, Hellgren & Bensch, 2007 and H. tartakovskyi ookinetes can be reported between 6 h and 3 dpe and sporozoites between 5 and 7 dpe, indicating asynchronous development ([2, 33, 38]; this study). In C. nubeculosus sporogony of H. homopalloris n. sp., H. noctuae, H. syrnii and H. tartakovskyi occurs at similar rates: oocysts can be seen 3 dpe until 5 dpe, and sporozoites about 7 dpe ([33, 34]; this study). Interestingly, the sporogony rate of Haemoproteus species inhabiting phylogenetically distantly related birds belong to different orders (Passeriformes and Strigiformes) is similar in same species of biting midges ([33, 34]; this study).
The presence of few oocysts in temporary preparations was likely due to the low parasitemia in the donor bird during this study (0.1%). Heavy Haemoproteus parasitemia is markedly virulent in blood-sucking insects, and such infections might kill them due to the damage by numerous migrating ookinetes [12]. When performing experimental infections with vectors it is important to use donor birds with light Haemoproteus sp. parasitemia (about 0.1%). If parasitemia is high (≥ 1%) in the blood meal, mortality is usually high in biting midges and other blood-sucking insects, resulting in rapid death of experimental groups [13]. Thus, donor birds should be carefully selected for experimental vector research. Interestingly, mainly light parasitemia (< 1%) usually is present in wild-caught birds [16, 19, 58, 73, 74], and such infections are particularly important for transmission. This explains the biological meaning of persistence of haemosporidian parasites in birds at stage of low parasitemia from the viewpoint of parasite transmission.
Conclusions
Haemoproteus homopalloris n. sp. (lineage hPHSIB2) is the third haemoproteid species reported in wood warblers. The new species belongs to the subgenus Parahaemoproteus based on the morphology of the sporogonic stages, susceptibility of Culicoides biting midges and phylogeny. The phylogenetic analysis identified a well-supported clade containing seven haemoproteids, including the new species, which possess pale staining macrogametocytes and develop exceptionally rapidly at ookinete stage; biological meaning of these characters remains unclear. Studies targeting mechanisms of sporogonic development of haemoproteids and other wildlife haemosporidian parasites remain uncommon and should be encouraged. Culicoides nubeculosus is an excellent experimental vector of H. homopalloris n. sp. and several other avian haemoproteids. This insect is relatively easy to rear, and it is recommended in laboratory experimental research.
References
Clark NJ, Clegg SM, Lima MR. A review of global diversity in avian haemosporidians (Plasmodium and Haemoproteus: Haemosporida): new insights from molecular data. Int J Parasitol. 2014;44:329–38.
Valkiūnas G. Avian Malaria Parasites and Other Haemosporidia. Boca Raton: CRC Press; 2005.
Bennett GF, Peirce MA, Ashford RW. Avian haematozoa: mortality and pathogenicity. J Nat Hist. 1993;27:993–1001.
Marzal A, De Lope F, Navarro C, Møller AP. Malarial parasites decrease reproductive success: an experimental study in a passerine bird. Oecologia. 2005;142:541–5.
Dunn JC, Goodman SJ, Benton TG, Hamer KC. Avian blood parasite infection during the non-breeding season: an overlooked issue in declining populations? BMC Ecol. 2013;13:30.
Cardona CJ, Ihejirika A, McClellan L. Haemoproteus lophortyx infection in bobwhite quail. Avian Dis. 2002;46:249–55.
Ferrell ST, Snowden K, Marlar AB, Garner M, Lung NP. Fatal hemoprotozoal infections in multiple avian species in a zoological park. J Zoo Wildl Med. 2007;38:309–16.
Donovan TA, Schrenzel M, Tucker TA, Pessier AP, Stalis IH. Hepatic hemorrhage, hemocoelom, and sudden death due to Haemoproteus infection in passerine birds: eleven cases. J Vet Diagn Invest. 2008;20:304–13.
Olias P, Wegelin M, Zenker W, Freter S, Gruber AD, Klopfleisch R. Avian malaria deaths in parrots, Europe. Emerg Infect Dis. 2011;17:950–2.
Cannell BL, Krasnec KV, Campbell K, Jones HI, Miller RD, Stephens N. The pathology and pathogenicity of a novel Haemoproteus spp. infection in wild little penguins (Eudyptula minor). Vet Parasitol. 2013;205:416.
Valkiūnas G, Iezhova TA. Exo-erythrocytic development of avian malaria and related haemosporidian parasites. Malar J. 2017;16:101.
Valkiūnas G, Kazlauskienė R, Bernotienė R, Bukauskaitė D, Palinauskas V, Iezhova TA. Haemoproteus infections (Haemosporida, Haemoproteidae) kill bird-biting mosquitoes. Parasitol Res. 2014;113:1011–8.
Bukauskaitė D, Bernotienė R, Iezhova TA, Valkiūnas G. Mechanisms of mortality in Culicoides biting midges due to Haemoproteus infection. Parasitology. 2016;143:1748–54.
Chagas CR, Guimarães L de O, Monteiro EF, Valkiūnas G, Katayama MV, Santos SV, et al. Hemosporidian parasites of free-living birds in the São Paulo Zoo, Brazil. Parasitol Res. 2016;115:1443–52.
Dimitrov D, Iezhova TA, Zehtindjiev P, Bobeva A, Ilieva M, Kirilova M, et al. Molecular characterisation of three avian haemoproteids (Haemosporida, Haemoproteidae), with the description of Haemoproteus (Parahaemoproteus) palloris n. sp. Syst Parasitol. 2016;93:431–49.
Matta NE, Pacheco MA, Escalante AA, Valkiūnas G, Ayerbe-Quiñones F, Acevedo-Cendales LD. Description and molecular characterization of Haemoproteus macrovacuolatus n. sp. (Haemosporida, Haemoproteidae), a morphologically unique blood parasite of black-bellied whistling duck (Dendrocygna autumnalis) from South America. Parasitol Res. 2014;113:2991–3000.
Mantilla JS, González AD, Lotta IA, Moens M, Pacheco MA, Escalante AA, et al. Haemoproteus erythrogravidus n. sp. (Haemosporida, Haemoproteidae): description and molecular characterization of a widespread blood parasite of birds in South America. Acta Trop. 2016;159:83–94.
Chagas CR, Valkiūnas G, de Oliveira Guimarães L, Monteiro EF, Guida FJ, Simões RF, et al. Diversity and distribution of avian malaria and related haemosporidian parasites in captive birds from a Brazilian megalopolis. Malar J. 2017;16:83.
Valkiūnas G, Pendl H, Olias P. New Haemoproteus parasite of parrots, with remarks on the virulence of haemoproteids in naive avian hosts. Acta Trop. 2017;176:256–62.
Ferreira-Junior FC, Dutra DA, Martins NRS, Valkiūnas G, Braga EM. Haemoproteus paraortalidum n. sp. in captive Black-fronted Piping-guans Aburria jacutinga (Galliformes, Cracidae): high prevalence in a population reintroduced into the wild. Acta Trop. 2018;188:93–100.
BirdLife International. Phylloscopus sibilatrix. The IUCN Red List of Threatened Species. 2016; https://doi.org/10.2305/IUCN.UK.2016-3.RLTS.T22715260A87668662.en. Accessed 22 May 2018.
Waldenström J, Bensch S, Kiboi S, Hasselquist D, Ottosson U. Cross-species infection of blood parasites between resident and migratory songbirds in Africa. Mol Ecol. 2002;11:1545–54.
Hellgren O, Waldenström J, Peréz-Tris J, Szöll E, Si O, Hasselquist D, et al. Detecting shifts of transmission areas in avian blood parasites: a phylogenetic approach. Mol Ecol. 2007;16:1281–90.
Bensch S, Hellgren O, Pérez-Tris J. MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol Ecol Resour. 2009;9:1353–8.
Shurulinkov P, Golemansky V. Haemoproteids (Haemosporida: Haemoproteidae) of wild birds in Bulgaria. Acta Protozool. 2002;41:359–74.
Ishtiaq F, Guillaumot L, Clegg SM, Phillimore AB, Black RA, Owens IPF, et al. Avian haematozoan parasites and their associations with mosquitoes across Southwest Pacific Islands. Mol Ecol. 2008;17:4545–55.
Kim KS, Tsuda Y, Sasaki T, Kobayashi M, Hirota Y. Mosquito blood-meal analysis for avian malaria study in wild bird communities: laboratory verification and application to Culex sasai (Diptera: Culicidae) collected in Tokyo, Japan. Parasitol Res. 2009;105:1351–7.
Njabo KY, Cornel AJ, Bonneaud C, Toffelmier E, Sehgal RN, Valkiūnas G, et al. Nonspecific patterns of vector, host and avian malaria parasite associations in a central African rainforest. Mol Ecol. 2011;20:1049–61.
Martínez-de la Puente J, Martínez J, Rivero de Aguilar J, Herrero J, Merino S. On the specificity of avian blood parasites: revealing specific and generalist relationships between haemosporidians and biting midges. Mol Ecol. 2011;20:3275–87.
Ferraguti M, Martinez-de la Puente J, Ruiz S, Soriguer R, Figuerola J. On the study of the transmission network of blood parasites from SW Spain: diversity of avian haemosporidians in the biting midge Culicoides circumscriptus and wild birds. Parasit Vectors. 2013;6:208.
Bobeva A, Zehtindjiev P, Ilieva M, Dimitrov D, Mathis A, Bensch S. Host preferences of ornithophilic biting midges of the genus Culicoides in the eastern Balkans. Med Vet Entomol. 2015;29:290–6.
Valkiūnas G, Kazlauskienė R, Bernotienė R, Palinauskas V, Iezhova TA. Abortive long-lasting sporogony of two Haemoproteus species (Haemosporida, Haemoproteidae) in the mosquito Ochlerotatus cantans, with perspectives on haemosporidian vector research. Parasitol Res. 2013;113:2159–69.
Bukauskaitė D, Žiegytė R, Palinauskas V, Iezhova TA, Dimitrov D, Ilgūnas M, et al. Biting midges (Culicoides, Diptera) transmit Haemoproteus parasites of owls: evidence from sporogony and molecular phylogeny. Parasit Vectors. 2015;8:303.
Žiegytė R, Bernotienė R, Palinauskas V, Valkiūnas G. Haemoproteus tartakovskyi (Haemoproteidae): complete sporogony in Culicoides nubeculosus (Ceratopogonidae), with implications for avian haemoproteid experimental research. Exp Parasitol. 2016;160:17–22.
Levin II, Valkiūnas G, Santiago-Alarcon D, Cruz LL, Iezhova TA, O'Brien SL, et al. Hippoboscid-transmitted Haemoproteus parasites (Haemosporida) infect Galapagos pelecaniform birds: evidence from molecular and morphological studies, with a description of Haemoproteus iwa. Int J Parasitol. 2011;41:1019–27.
Santiago-Alarcon D, Palinauskas V, Schaefer HM. Diptera vectors of avian haemosporidian parasites: untangling parasite life cycles and their taxonomy. Biol Rev Camb Philos Soc. 2012;87:928–64.
Bobeva A, Zehtindjiev P, Bensch S, Radrova J. A survey of biting midges of the genus Culicoides Latreille, 1809 (Diptera: Ceratopogonidae) in NE Bulgaria, with respect to transmission of avian haemosporidians. Acta Parasitol. 2013;58:585–91.
Žiegytė R, Palinauskas V, Bernotienė R, Iezhova TA, Valkiūnas G. Haemoproteus minutus and Haemoproteus belopolskyi (Haemoproteidae): complete sporogony in the biting midge Culicoides impunctatus (Ceratopogonidae), with implications on epidemiology of haemoproteosis. Exp Parasitol. 2014;145:74–9.
Atkinson CT. Haemoproteus. In: Atkinson CT, Thomas NJ, Hunter BC, editors. Parasitic Diseases of Wild Birds. Ames: Wiley-Blackwell; 2008. p. 13–35.
Nayduch D, Cohnstaedt LW, Saski C, Lawson D, Kersey P, Fife M, et al. Studying Culicoides vectors of BTV in the post-genomic era: resources, bottlenecks to progress and future directions. Virus Res. 2014;182:43–9.
Boorman J. The maintenance of laboratory colonies of Culicoides variipenniss (Coq.), C. nubeculosus (Mg.) and C. riethi Kieff. (Diptera, Ceratopogonidae). Bull Entomol Res. 1974;64:371–7.
Pages N, Breard E, Urien C, Talavera S, Viarouge C, Lorca-Oro C, et al. Culicoides midge bites modulate the host response and impact on bluetongue virus infection in sheep. PLoS One. 2014;9:e83683.
Miltgen F, Landau I, Ratanaworabhan N, Yenbutra S. Parahaemoproteus desseri n. sp.; Gametogonie et shizogonie chez I’hote naturel: Psittacula roseate de Thailande, et sporogonie experimentale chez Culicoides nubeculosus. Ann Parasitol Hum Comp. 1981;56:123–30.
Valkiūnas G, Iezhova TA, Križanauskienė A, Palinauskas V, Sehgal RNM, Bensch S. A comparative analysis of microscopy and PCR-based detection methods for blood parasites. J Parasitol. 2008;94:1395–401.
Godfrey RD, Fedynich AM, Pence DB. Quantification of hematozoa in blood smears. J Wildl Dis. 1987;23:558–65.
Richardson DS, Jury FL, Blaakmeer K, Komdeur J, Burke T. Parentage assignment and extra-group paternity in a cooperative breeder: the Seychelles warbler (Acrocephalus sechellensis). Mol Ecol. 2001;10:2263–73.
Bensch S, Stjernman M, Hasselquist D, Ostman O, Hansson B, Westerdahl H, et al. Host specificity in avian blood parasites: a study of Plasmodium and Haemoproteus mitochondrial DNA amplified from birds. Proc Biol Sci. 2000;267:1583–9.
Hellgren O, Waldenström J, Bensch S. A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium and Haemoproteus from avian blood. J Parasitol. 2004;90:797–802.
Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–8.
Ronquist F, Heulsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4.
Nylander JAA. MrModeltest v2. Program distributed by the author. Software available at https://github.com/nylander/MrModeltest2. Accessed 10 Apr 2018.
Rambaut A. FigTree: Tree Figure Drawing Tool Version 1.4.0. 2006–2012. Institute of Evolutionary Biology, University of Edinburgh. http://tree.bio.ed.ac.uk/software/figtree/.
Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33:1870–4.
ICZN. International Commission on Zoological Nomenclature: Amendment of articles 8, 9, 10, 21 and 78 of the International Code of Zoological Nomenclature to expand and refine methods of publication. Bull Zool Nomencl. 2012;69:161–9.
Drovetski SV, Aghayan SA, Mata VA, Lopes RJ, Mode NA, Harvey JA, et al. Does the niche breadth or trade-off hypothesis explain the abundance-occupancy relationship in avian haemosporidia? Mol Ecol. 2014;23:3322–9.
Valkiūnas G, Iezhova TA, Loiseau C, Chasar A, Smith TB, Sehgal RNM. New species of haemosporidian parasites (Haemosporida) from African rainforest birds, with remarks on their classification. Parasitol Res. 2008;103:1213–28.
Križanauskienė A, Pérez-Tris J, Palinauskas V, Hellgren O, Bensch S, Valkiūnas G. Molecular phylogenetic and morphological analysis of haemosporidian parasites (Haemosporida) in a naturally infected European songbird, the blackcap Sylvia atricapilla, with description of Haemoproteus pallidulus sp. nov. Parasitology. 2010;137:217–27.
Dimitrov D, Zehtindjiev P, Bensch S, Ilieva M, Iezhova TA, Valkiūnas G. Two new species of Haemoproteus Kruse, 1890 (Haemosporida, Haemoproteidae) from European birds, with emphasis on DNA barcoding for detection of haemosporidians in wildlife. Syst Parasitol. 2014;87:135–51.
Valkiūnas G, Iezhova TA. Keys to the avian malaria parasites. Malar J. 2018;17:212.
Hellgren O, Krizanauskiene A, Valkiūnas G, Bensch S. Diversity and phylogeny of mitochondrial cytochrome B lineages from six morphospecies of avian Haemoproteus (Haemosporida: Haemoproteidae). J Parasitol. 2007;93:889–96.
Valkiūnas G, Križanauskienė A, Iezhova TA, Hellgren O, Bensch S. Molecular phylogenetic analysis of circumnuclear hemoproteids (Haemosporida: Haemoproteidae) of sylvid birds, with a description of Haemoproteus parabelopolskyi sp. nov. J Parasitol. 2007;93:680–7.
Levin II, Valkiūnas G, Iezhova TA, O'Brien SL, Parker PG. Novel Haemoproteus species (Haemosporida: Haemoproteidae) from the swallow-tailed gull (Laridae), with remarks on the host range of hippoboscid-transmitted avian hemoproteids. J Parasitol. 2012;98:847–54.
Palinauskas V, Iezhova TA, Križanauskienė A, Markovets MY, Bensch S, Valkiūnas G. Molecular characterization and distribution of Haemoproteus minutus (Haemosporida, Haemoproteidae): a pathogenic avian parasite. Parasitol Int. 2013;62:358–63.
Valkiūnas G, Iezhova TA, Shapoval AP. High prevalence of blood parasites in hawfinch Coccothraustes coccothraustes. J Nat Hist. 2003;37:2647–52.
Valkiūnas G, Bensch S, Iezhova TA, Križanauskienė A, Hellgren O, Bolshakov CV. Nested cytochrome b polymerase chain reaction diagnostics underestimate mixed infections of avian blood haemosporidian parasites: microscopy is still essential. J Parasitol. 2006;92:418–22.
Merino S, Moreno J, Vásquez RA, Martínez J, Sánchez-Monsálvez I, Estades CF, et al. Haematozoa in forest birds from southern Chile: latitudinal gradients in prevalence and parasite lineage richness. Austral Ecol. 2008;33:329–40.
Marzal A, Ricklefs RE, Valkiūnas G, Albayrak T, Arriero E, Bonneaud C, et al. Diversity, loss, and gain of malaria parasites in a globally invasive bird. PLoS One. 2011;6:e21905.
Chagas CR, Valkiūnas G, Nery CV, Henrique PC, Gonzalez IH, Monteiro EF, et al. Plasmodium (Novyella) nucleophilum from an Egyptian goose in São Paulo Zoo, Brazil: microscopic confirmation and molecular characterization. Int J Parasitol Parasites Wildl. 2013;2:286–91.
Palinauskas V, Valkiūnas G, Bolshakov CV, Bensch S. Plasmodium relictum (lineage SGS1) and Plasmodium ashfordi (lineage GRW2): the effects of the co-infection on experimentally infected passerine birds. Exp Parasitol. 2011;127:527–33.
Martinez J, Martinez-De La Puente J, Herrero J, Del Cerro S, Lobato E, Rivero-De Aguilar J, et al. A restriction site to differentiate Plasmodium and Haemoproteus infections in birds: on the inefficiency of general primers for detection of mixed infections. Parasitology. 2009;136:713–22.
Bernotienė R, Palinauskas V, Iezhova T, Murauskaitė D, Valkiūnas G. Avian haemosporidian parasites (Haemosporida): a comparative analysis of different polymerase chain reaction assays in detection of mixed infections. Exp Parasitol. 2016;163:31–7.
Pacheco MA, Cepeda AS, Bernotienė R, Lotta IA, Matta NE, Valkiūnas G, et al. Primers targeting mitochondrial genes of avian haemosporidians: PCR detection and differential DNA amplification of parasites belonging to different genera. Int J Parasitol. 2018;48:657–70.
Dimitrov D, Ilieva M, Ivanova K, Brlík V, Zehtindjiev P. Detecting local transmission of avian malaria and related haemosporidian parasites (Apicomlexa, Haemosporida) at a Special Protection Area of Natura 2000 network. Parasitol Res. 2018;117:2187–99.
Ivanova K, Zehtindjiev P, Mariaux J, Dimitrov D, Georgiev BB. Avian haemosporidians from rain forests in Madagascar: molecular and morphological data of the genera Plasmodium, Haemoproteus and Leucocytozoon. Infect Genet Evol. 2018;58:115–24.
Acknowledgments
We thank V. Jusys and V. Eigirdas (Ventės Ragas Ornithological Station, Lithuania) for support during field work and R. Bernotienė, for assistance in the laboratory. Institute for Animal Health, Pirbright Laboratory for providing Culicoides nubeculosus for establishing our colony as part of a grant funded by the Biotechnology and Biological Sciences Research Council (BBS/E/I/00001701), UK.
Funding
This study was funded by the Research Council of Lithuania (nr. DOTSUT-137-09.3.3-LMT-K-712-02-0004) and also supported by the Open Access to research infrastructure of the Nature Research Centre under Lithuanian open access network initiative.
Availability of data and materials
The data supporting the findings of this study are included within the article. Representative preparations of blood (accession numbers 49021 NS and 49022 NS) and vector stages (49023 NS and 49024 NS) were deposited in the Nature Research Centre, Vilnius, Lithuania. Parahapantotype of the new species was deposited in the Queensland Museum, Brisbane, Australia (accession number G466204). The sequences was deposited in the GenBank database under the accession number MH513601.
Author information
Authors and Affiliations
Contributions
Experimental conception and design: GV and CRFC. Biting midge experiments, dissection and laboratory maintenance: DB. Donor bird collection and testing: GV, MI and TI. Morphological analysis: CRFC, GV and TAI. Phylogenetic analysis and its discussion: CRFC and GV. Paper writing: CRFC and GV. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
The experiments described herein comply with the current laws of Lithuania, have been performed by licenced researchers and were approved by the Lithuania and Environmental Protection Agency, Vilnius (2017-04-26, no. 23). None of the experimental birds suffered apparent injury during sampling and all birds were released.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Chagas, C.R.F., Bukauskaitė, D., Ilgūnas, M. et al. A new blood parasite of leaf warblers: molecular characterization, phylogenetic relationships, description and identification of vectors. Parasites Vectors 11, 538 (2018). https://doi.org/10.1186/s13071-018-3109-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-018-3109-9