Abstract
Background
Haemoproteus parasites are widespread, and several species cause diseases both in birds and blood-sucking insects. These pathogens are transmitted by dipterans belonging to the Ceratopogonidae and Hippoboscidae, however certain vector species remain unknown for the majority of Haemoproteus spp. Owls are often infected by Haemoproteus parasites, but experimental studies on vectors of these infections are lacking. The aim of this study was to investigate sporogonic development of two widespread Haemoproteus parasites of owls, H. noctuae and H. syrnii in experimentally infected biting midges Culicoides impunctatus and Culicoides nubeculosus. We also followed in vitro sporogonic development of these infections and determined their phylogenetic relationships with Haemoproteus spp., for which vectors have been identified.
Methods
Wild-caught C. impunctatus and laboratory reared C. nubeculosus were infected experimentally by allowing them to take blood meals on one individual long-eared owl (Asio otus) and one tawny owl (Strix aluco) harbouring mature gametocytes of H. noctuae (lineage hCIRCUM01) and H. syrnii (hCULCIB01), respectively. The engorged insects were maintained in the laboratory at 16–18 °C, and dissected at intervals in order to follow the development of ookinetes, oocysts and sporozoites. We also observed in vitro development of sexual stages of both parasites by exposure of infected blood to air. The parasite lineages were determined by polymerase chain reaction-based methods. Bayesian phylogeny was constructed in order to determine the relationships of owl parasites with other avian Haemoproteus spp., for which vectors have been identified.
Results
Both H. noctuae and H. syrnii completed sporogony in C. nubeculosus, and H. noctuae completed sporogony in C. impunctatus. Ookinetes, oocysts and sporozoites of these parasites were reported and described. Gametes and ookinetes of both species readily developed in vitro. In accordance with sporogony data, the phylogenetic analysis placed both parasite lineages in a clade of Culicoides spp.-transmitted avian Haemoproteus (Parahaemoproteus) spp.
Conclusions
Culicoides nubeculosus and C. impunctatus are vectors of H. noctuae and H. syrnii. Phylogenies based on cytochrome b gene indicate parasite-vector relationships, and we recommend using them in predicting possible parasite-vector relationships and planning research on avian Haemoproteus spp. vectors in wildlife.
Similar content being viewed by others
Background
Haemoproteus parasites (Haemosporida, Haemoproteidae) are distributed worldwide, and several species cause diseases, sometimes severe, both in birds and blood-sucking insects [1–11]. This genus of haemosporidians contains subgenera Parahaemoproteus and Haemoproteus, where species are transmitted by blood-sucking insects of the Ceratopogonidae and Hippoboscidae, respectively [12–16]. In spite of numerous recent publications on avian haemosporidians [17–22], a few studies address Haemoproteus spp. vectors and transmission of these pathogens [14, 23–26], and experimental studies on these issues are few [5, 16, 27].
Haemoproteus parasites are widespread in owls worldwide, with prevalence of infection exceeding 50 % in many owl populations [28–31]. Pathogenicity of Haemoproteus infections in owls has been insufficiently investigated [5], but there is some evidence that these infections can be harmful. The co-infection of Leucocytozoon danilewskyi and Haemoproteus noctuae is likely responsible for mortality in snowy owl (Nyctea scandiaca), probably due to high parasitaemia [32]. However, vectors of this infection and other owl haemoproteids remain unidentified. A recent study based on observation of Haemoproteus syrnii in single naturally infected louse fly Ornithomyia sp., speculated that this widespread parasite of owls might be transmitted by hippoboscid flies [33]. However, traditional classifications placed H. syrnii and other haemoproteids of owls to the subgenus Parahaemoproteus, suggesting that they are transmitted by Culicoides spp. [5, 12]. Because experimental studies on vectors of owl Haemoproteus parasites are lacking, the aim of this study was to investigate sporogony of H. noctuae and H. syrnii in experimentally infected biting midges Culicoides impunctatus and C. nubeculosus. These biting midges are widespread in Europe and willingly take blood meals on birds [34]. Additionally, development of sexual stages of both these parasites was initiated in vitro. We also determined phylogenetic relationships among Haemoproteus spp., for which vectors have been identified, and discussed this information in regard to the possible perspectives in vector research of haemoproteids.
Methods
Study site, selection of experimental birds and collection of blood samples
Experiments were carried out at the Biological Station of the Zoological Institute of the Russian Academy of Sciences on the Curonian Spit in the Baltic Sea (55°05' N, 20°44' E) between 9 and 24 June in 2014.
In May 2014, one long-eared owl (Asio otus) and one common crossbill (Loxia curvirostra) were captured with big Rybachy traps [35] at the study site. Three tawny owls (Strix aluco) were examined for blood parasites in Kaliningrad Zoo, and we selected one bird for this experimental study. The experimental tawny owls originally came to the zoo from Kaliningrad District, Russia. All birds were kept outside in cages, which were protected from the penetration of biting midges and other blood-sucking insects by covers made of fine-mesh bolting silk. The birds were maintained at natural light–dark photoperiod (L/D) 17:7 h. The owls were fed with wild-caught mice and naturally died wild birds; they were held for approximately two weeks (the adaptation period) before they were used as donors of gametocytes in experiments. Both owls and the crossbill survived to the end of this study and were released after experimental work at the study site, which is a natural breeding area for these bird species.
Approximately 30 μl of blood was collected with microcapillaries by punching the brachial vein. One drop was used to make three blood films. The smears were air-dried within 5 – 15 s after their preparation, fixed in absolute methanol and stained with 10 % Giemsa solution, as described by Valkiūnas [5]. The blood smears were examined microscopically to identify parasite species and to control for the possible presence of natural co-infections with other haemosporidian parasites. Remaining blood was stored in non-lysis SET buffer (0.05 M Tris, 0.15 M NaCl, 0.5 M EDTA, pH 8.0) for molecular analysis. The samples were held at ambient temperature in the field and later at −20 °C in the laboratory.
Blood samples from donor birds were examined by polymerase chain reaction (PCR)-based methods. Amplifications were sequenced and cytochrome b (cyt b) lineages of Haemoproteus parasites were determined in the laboratory (see description below). One long-eared owl and one tawny owl with single infections of H. noctuae (cyt b lineage hCIRCUM01, gametocytaemia of 0.07 %) and H. syrnii (CULKIB01, gametocytaemia of 0.03 %), as determined both by microscopic examination and PCR-based testing, were used as donors of gametocytes to infect biting midges and to carry out in vitro experiments. Parasite species were identified according to Valkiūnas [5]. One uninfected juvenile common crossbill (Loxia curvirostra) was used to feed a control group of flies.
Experimental design
Experimental infections of biting midges were performed in two ways. First, wild biting midges C. impunctatus were exposed to Haemoproteus infections near the Lake Chaika, located close to the village Rybachy, where density of C. impunctatus was high [36]. Second, we used the laboratory reared C. nubeculosus in experiments. Below, we describe these methods in more detail.
Wild-caught C. impunctatus biting midges were infected by allowing them to take blood meals on two infected owls and one uninfected crossbill (control), as described by Valkiūnas [5]. Briefly, the birds were held in the hands covered by rubber gloves. Biting midges were allowed to feed naturally on infected birds between 10 pm and 12 pm. The birds were exposed to bites for approximately 1.5–2 h depending on density of biting midges at the study site. This biting midge willingly takes blood meals on the feather-free region of body, and the insects were particularly often observed close to eyes (Fig. 1a). When several females began taking blood meals on a bird’s head, the head with feeding insects was carefully placed into an unzipped insect cage (approximately 12 × 12 × 12 cm) made of fine-mesh bolting silk. The engorged females fly off after the blood meals. The cage with engorged biting midges was then closed using a zipper. Up to 20 females were allowed to take blood meals on each bird daily. Cages with engorged flies were transported to the laboratory. To feed the biting midges, several pads of cotton-wool moistened with 10 % saccharose solution were placed on the top of each cage. Thirty-four infected C. impunctatus insects were used for dissection.
Experimental infection (a, c, d) and dissection (b) of the biting midges Culicoides impunctatus (a, b) and Culicoides nubeculosus (c, d) for detection of sporogonic stages of owl Haemoproteus spp. a – engorged biting midge C. impunctatus (long arrow) taking blood meal close to an eye of the long-eared owl Asio otus; b – dissection of C. impunctatus, short arrow indicates a dissecting needle; c – a hand-held cardboard box with a colony of C. nubeculosus inside, arrowheads indicate the midges sitting on fine-mesh bolting silk covering one side of the box; d – experimental infection of the colony of C. nubeculosus midges by means of adherence of a box with flies to skin of the tawny owl Strix aluco, triangle arrowhead indicates the box with flies
Culicoides nubeculosus biting midges were reared in the laboratory according to Boorman [37]. Each insect colony was maintained in small (approximately 5 cm in diameter) cardboard boxes covered by fine mesh bolting silk (Fig. 1c). The laboratory reared biting midges were delivered to the Biological Station and exposed by gentle pressing of a box with flies to a feather-free area on pectoral muscles of owls (Fig. 1d). Biting midges willingly took blood meals via fine-mesh bolting silk, and the majority of females were fully engorged approximately 30 min after the exposure. Twenty-four infected C. nubeculosus insects were used for dissection.
Both wild caught C. impunctatus and laboratory reared C. nubeculosus were held in the same room and conditions, i. e. 16–18 °C, 70 ± 5 % relative humidity and L/D photoperiod of 17:7 h. Biting midges were fed with 10 % saccharose solution; pads of cotton-wool were moistened in this solution and placed on the top of each insect cage daily.
In order to determine prevalence of possible natural Haemoproteus infection in wild-caught C. nubeculosus, 200 females were collected at the study site and tested by PCR-based methods (see description below). DNA was extracted from 40 pools of biting midges, each containing 5 flies.
Dissection of biting midges and making preparations of parasites
Before dissection, biting midges were identified morphologically according to Gutsevich [38]. Insects were dissected at intervals in order to follow the development of ookinetes, oocysts and sporozoites. Engorged females were anesthetized by putting them into a tube with a cotton pad wetted in 96 % ethanol. Then, each insect was placed in a drop of 0.9 % normal saline. Midguts and salivary glands were extracted using dissecting needles (Fig. 1b), which were disinfected in fire to avoid contaminations after each dissection. We examined midgut contents for ookinetes 0.5–1 day post exposure (dpe), midgut wall for oocysts 3–8 dpe, and salivary glands for sporozoites 6–10 dpe.
Parasite preparations were prepared according to Valkiūnas [5]. Briefly, the midguts and salivary glands were extracted, gently crashed to prepare small smears, air dried, fixed with methanol and stained with Giemsa. Midgut preparations were stained in the same way as blood smears, while 4 % staining solution was used to stain preparations of salivary glands. Oocysts were first visualised by adding a minute drop of 2 % mercurochrome solution on freshly prepared midgut preparation, which was then covered with a coverslip. That simplifies the search of tiny Parahaemoproteus parasite oocysts. Oocyst-infected midguts were fixed in 10 % formalin solution for 24 h; formalin was then replaced with 70 % ethanol. After 6 h, the midgut preparations were washed with distilled water, stained with Ehrlich’s hematoxylin for 10 min, steeped in water containing a pinch of sodium bicarbonate and differentiated with acid ethanol both for 5 min and again steeped in water with sodium bicarbonate. Then, each preparation was dehydratated with 70 % and 96 % ethanol, cleared by putting a drop of clove oil and xylene over the preparation, and finally mounted in Canada balsam and covered with a cover slip. After dissection, all residual parts of insects were placed in 96 % ethanol to confirm the presence of parasite lineages by PCR-based methods (see description below).
In vitro sporogonic development
Approximately 300 μl of blood was taken from the brachial vein of the infected owls. The blood was immediately placed in an Eppendorf tube and diluted with 3.7 % solution of sodium citrate (pH 8.5) in ratio 1 part of solution to 4 parts of blood. The work was performed at 20 °C ± 1 °C. To follow in vitro development of the parasites, smears were prepared 12 h after exposure to air. They were air-dried, fixed in methanol and stained with Giemsa, as described for blood films.
Microscopic examinations of preparations and parasite morphology
All preparations were examined with Olympus BX-43 light microscope equipped with Olympus SZX2-FOF digital camera and imaging software QCapture Pro 6.0, Image-Pro plius (Tokyo, Japan). Blood films were examined for 15–20 min at low magnification (×400), and then at least 100 fields were studied at high magnification (×1000). Intensity of parasitaemia was estimated each day before exposure of biting midges; it was determined as a percentage by actual counting of the number of mature gametocytes (Fig. 2a–d) per 1000 red blood cells. All vector preparations were first examined at low magnification (×100, ×600) and then at high magnification (×1000). The statistical analyses were carried out using the “Statistica 7” package. Student’s t-test for independent samples was used to determine statistical significance between mean linear parameters of parasites. A P value of 0.05 or less was considered significant. Representative preparations of blood stages (accession nos. 418836–48838 NS) and vector stages in vitro (48840–48844 NS) and in vivo (48845–48856 NS) were deposited in Nature Research Centre, Vilnius, Lithuania. Two voucher preparations of blood stages were deposited in the Queensland Museum, Queensland, Australia (accessions G465772, G465773).
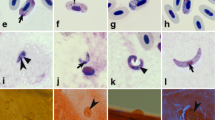
Mature gametocytes (a–d) and sporogonic stages (e–j) of Haemoproteus noctuae (lineage hCIRCUM01) in Culicoides nubeculosus (e, i) and Culicoides impunctatus (g), and Haemoproteus syrnii (hCULCIB01) in C. nubeculosus (f, h, j). Macrogametocytes (a, b) and microgametocytes (c, d) of H. noctuae (a, c) and H. syrnii (b, d) in peripheral blood of donor birds before experimental infection of biting midges. Ookinetes (e, f), oocyst (h) and sporozoites (g, i, j) are shown. Long simple arrows – nuclei of parasites, simple arrowheads – pigment granules, triangle arrowheads – volutin granules, short arrow – oocyst. Scale bar = 10 μm
Polymerase chain reaction and sequencing
Total DNA was extracted from all samples using ammonium acetate extraction method [39]. For genetic analysis, we used a nested PCR protocol [40, 41]. Amplification of the cyt b gene was done using two pairs of initial primers, HaemNFI and HaemNR3 for detection of Haemoproteus, Plasmodium and Leucocytozoon species. For the second PCR, we used primers HaemF and HaemR2, which are specific to Haemoproteus and Plasmodium parasites. All samples were evaluated by running 1.5 μl of PCR product on a 2 % agarose gel. One negative control (nuclease-free water) and one positive control (an infected sample, which was positive by microscopic examination of blood films) were used per every 14 samples. No cases of false positive or negative samples were reported.
To support species identification of wild-caught biting midges, we used the insect-specific primers LCO149 and HCO2198, which amplify a fragment of cytochrome oxidase subunit I of mitochondrial DNA [42].
Fragments of DNA from the PCR positive samples were sequenced from the 3′ and 5′ ends. The genetic analyzer “Basic Local Alignment Search Tool” (National Centre of Biotechnology Information website: http//www.ncbi.nlm.nih.gov/BLAST) was used to determine lineages of detected DNA sequences. Sequences were edited and aligned using BioEdit [43] and deposited in GenBank (accessions KP794611 and KP794612).
Phylogenetic analysis
To determine phylogenetic relationships of H. syrnii and H. noctuae with other avian haemoproteids, for which vectors have been identified, we constructed a phylogenetic tree using 21 sequences of the mitochondrial cyt b gene of Haemoproteus spp. and three sequences of Plasmodium spp.; each sequence was of 479 bp. The tree was created using Bayesian phylogenetics as implemented in mrBayes version 3.1 [44]. Best-fit model of evolution (GTR + I + G) was selected by software Modeltest 3.7 [45]. We ran two independent analyses with a sample frequency of every 100th generation over 12 million generations. For construction of the majority consensus tree, 25 % of the initial trees in each run were discarded as burn in periods. We visualized the tree using the software Tree View 1.6.6. (available from <http://evolution.genetics.washington.edu/phylip/software.html>). The sequence divergence between the different lineages was calculated with the use of a Jukes-Cantor model of substitution, with all substitutions weighted equally, implemented in Molecular Evolutionary Genetics Analysis (MEGA) software, version 4.0 [46].
Ethical statement
The experiments described herein comply with the current laws of Lithuania and Russia. Experimental procedures were approved by the International Research Co-operation Agreement between the Biological Station Rybachy of the Zoological Institute of the Russian Academy of Sciences and Institute of Ecology of Nature Research Centre (25-05-2010). Kaliningrad Zoo provided one naturally infected tawny owl for experimental research according to the ethical approval of 05-06-2014. All efforts were made to minimize handling time and potential suffering of birds. None of the experimental birds suffered apparent injury during experiments and were released after experiments.
Results
Common crossbill used for control was parasite negative both by PCR and microscopic examination. Morphological examination and PCR-based testing showed that all experimental wild-caught flies belonged to C. impunctatus. According to PCR-based analysis, no natural infection was found in wild-caught biting midges. The lack of infected insects among wild-caught females is not surprising because 1) the natural infection of biting midges with hemoproteids usually is light (<1 %, see Valkiūnas [5]), and we carried out experiments in the beginning of the season of their activity when females likely took the first blood meal, so a probability to get natural infection was light. Parasites were also not detected in control flies both by PCR-based and microscopic examination methods.
In accordance to microscopic observation, the PCR and sequencing confirmed the presence of corresponding parasite lineages hCIRCUM01 and hCULCIB01 in experimentally infected biting midges.
Sporogony of H. noctuae and H. syrnii in biting midges
Sporogonic development of the parasites was observed in all dissected infected insects. Ookinetes, oocysts and sporozoites of H. noctuae were seen both in experimentally infected wild-caught C. impunctatus and laboratory reared C. nubeculosus biting midges, indicating completed sporogony (Fig. 2e–j). Haemoproteus syrnii completed sporogony in C. nubeculosus, but only ookinetes and oocyst were found in C. impunctatus. Sporozoites of H. syrnii were not seen in salivary gland preparations of the latter biting midge probably due to the light infection. Additionally, only 2 exposed flies survived and were dissected 7 dpe, and sporozoites might not reach salivary glands or were few and difficult to find because of the small sample and short observation period.
Ookinetes of H. noctuae were seen 12 h post exposure both in C. impunctatus and C. nubeculosus (Fig. 2e). Ookinetes of H. syrnii were reported in C. nubeculosus 14 h post exposure (hpe) (Fig. 2f). Ookinetes of both species were elongated worm-like bodies possessing prominent, slightly off centre located nuclei and large ‘vacuoles’. Residual pigment granules were clamped at the distal ends of the parasites (Fig. 2e, f). In C. impunctatus, ookinetes of H. noctuae measured (n = 20) 17.0–24.5 (on average 20.7 ± 1.9) μm in length, 1.8–2.9 (2.4 ± 0.3) μm in width, 32.3–54.5 (41.2 ± 5.5) μm in area, and 3.1–8.0 (5.5 ± 1.6) μm in nucleus area. A few ookinetes of H. syrnii were seen, and they were not measured.
Oocysts of H. noctuae and H. syrnii were seen in fresh preparations both in C. impunctatus and C. nubeculosus biting midges 4–5 dpi, but only oocysts of H. syrnii (Fig. 2h) were observed and measured in permanent preparations. Oocysts of Hamoproteus parasites are tiny and difficult to observe and calculate on midguts; between 1 and 30 oocysts were observed in different oocyst preparations. Flattened oocysts of H. syrnii measured (n = 21) 4.0–8.1 (5.4 ± 0.9) μm in minimum diameter, 5.1–8.3 (6.1 ± 0.8) μm in maximum diameter, and 19.7–60.4 (27.7 ± 9.7) μm, in area.
Sporozoites of H. noctuae (Fig. 2g, i) and H. syrnii (Fig. 2j) were reported in salivary gland preparations 7–9 dpi, indicating that these biting midges are likely vectors of these parasites.
Morphologically identical sporozoites of H. noctuae developed both C. impunctatus and C. nubeculosus; they were fusiform bodies with off centre located nuclei and approximately equally pointed ends (Fig. 2g, i). There was no significant difference in the length, widths, area or area of nuclei in H. noctuae sporozoites (Table 1), which developed into different species of biting midges (P > 0.05 for all these features). Sporozoites of H. syrnii were also fusiform bodies with approximately equally pointed ends, but nuclei were located centrally (Fig. 2j). Additionally, sporozoites of H. syrnii were significantly shorter and smaller in area than those of H. noctuae (P < 0.05 for both these features). Based on these characteristics, sporozoites of H. noctuae and H. syrnii can be readily distinguished from each other (Table 1).
In vitro development of Haemoproteus noctuae and H. syrnii
Microgametes, macrogametes and ookinetes of H. noctuae and H. syrnii developed in vitro and were observed 12 h after exposure of mature gametocytes to air (Fig. 3a–f). Macrogametes of both species possess prominent pigment granules (Fig. 3a, d). Volutin, which was inherited from macrogametocytes (Fig. 2b), is visible in H. syrnii macrogametes (Fig. 3d). Microgametes of both parasites were thread-like bodies possessing readily visible nuclei (Fig. 3b, e). Ookinetes of H. noctuae and H. syrnii were worm-like bodies possessing prominent nuclei, ‘vacuoles’ and pigment granules (Fig. 3c, f). Volutin granules were invisible in ookinetes of H. syrnii (Fig. 3f). Both H. noctuae and H. syrnii ookinetes, which developed in vitro, were morphologically similar to those developed in vivo (compare Fig. 2e, f with Fig. 3c, f).
In vitro mature gametes and ookinetes of Haemoproteus noctuae (lineage hCIRCUM01, a–c) and Haemoproteus syrnii (hCULCIB01, d–f). Macrogametes (a, d), microgametes (b, e) and ookinetes (c, f) are shown. Long simple arrows – nuclei of parasites, simple arrowheads – pigment granules, triangle arrowheads – volutin granules. Scale bar = 10 μm
Phylogenetic analysis
Phylogenetic analysis placed all Culicoides spp.-transmitted avian haemoproteids in a well-supported clade A (Fig. 4), which contains Parahaemoproteus parasites. One H. noctuae lineage and five H. syrnii lineages, including those used in this study (Fig. 4, clade C), appeared in the clade A. That is in accordance with our sporogony study, which showed complete sporogonic development of these parasites in Culicoides biting midges (Fig. 2e–j). Lineages of hippoboscid-transmitted parasites of the subgenus Haemoproteus were placed in a separate well-supported clade B, which is the sister to clade A (Fig. 4).
Bayesian phylogeny of 16 Haemoproteus (Parahaemoproteus) spp., 5 H. (Haemoproteus) spp. and 3 Plasmodium spp. based on cytochrome b gene sequences of 479 bp lengths. One Leucocytozoon sp. sequence was used as outgroup. Nodal support values indicate Bayesian posterior probabilities. The scale bar shows the expected substitutions per site. Parasite lineage codes are given according to MalAvi database [17]; they are followed by GenBank accession numbers of sequences and parasite species names. Vertical bars (a–c) indicate groups of closely related species belonging to the subgenus Parahaemoproteus (a) and Haemoproteus (b); lineages of owl parasites are marked by bar C. Parasite lineages used in this study are given in bold font
Discussion
There are several key results of this study. First, we add H. noctuae and H. syrnii to the list of Culicoides spp.-transmitted haemoproteids, supporting their belonging to the subgenus Parahaemoproteus [5, 12, 13]. Both these parasites completed sporogony in Culicoides biting midges, with sporozoites reported in salivary glands (Fig. 2g, i, j), indicating that these flies likely are natural vectors, as has been reported for Parahaemoproteus species [2, 5, 16]. Several other Parahaemoproteus parasites have been shown experimentally to complete sporogony in Culicoides spp. These are H. danilewskii [47, 48], H. dolniki [5], H. handai [1], H. mansoni [27], H. nettionis [49], and H. velans [50]. However, molecular characterisation of these avian haemoproteids has not been developed and cyt b sequence information is absent, thus they cannot be included in the phylogenetic analysis (Fig. 4).
Second, the phylogenetic analysis placed cyt b lineages both of H. noctuae and H. syrnii in the clade A (Fig. 4). This clade contains nine species of avian Parahaemoproteus spp., for which molecular characterization has been developed and sporogony investigated experimentally. These parasites infect passeriform and strigiform birds and complete sporogony in Culicoides biting midges [5, 16]. This phylogenetic study (Fig. 4) confirms results of our experiments. In other words, vector studies and phylogenetic analysis based on cyt b gene complement each other [5, 16, 51, this study], indicating that the genetic differences in cyt b gene between Parahaemoproteus and Haemoproteus parasites likely reflect differences in their sporogonic stages. Mainly, species of the subgenus Parahaemoproteus are characterised by small oocysts (<20 μm in diameter) that possess one germinative centre, a relatively small number of sporozoites in mature oocysts (<100), sporozoites that are usually pointed at both ends, transmitted by biting midges belonging to the Ceratopogonidae, and some other features [5, 13, 52]. Haemoproteids, which are transmitted by hippoboscid flies (Hippoboscidae) belong to the subgenus Haemoproteus; they are characterized by large oocysts (>20 μm in diameter) that possess numerous germinative centres, relatively many sporozoites in mature oocysts (>500 sporozoites) that are usually blunt at one end and pointed in the other, and some other features [5, 13, 52]. Based on obtained results (Figs. 2, 3 and 4), we predict that all cyt b sequences of haemoproteids transmitted by biting midges will be grouped with the closely related lineages of parasites of the clade A (Fig. 4), and the lineages of hippoboscid transmitted parasites will be clustered with lineages of the clade B (Fig. 4). In other words, the phylogenetic analyses of cyt b sequences likely provide opportunities to predict groups of vectors (species of the Ceratopogonidae or Hippoboscidae), which are involved in transmission of avian haemoproteids. Determination of haemosporidian vectors is a time-consuming process, which requires more parasitology research, including infection and dissection of blood-sucking insects and search for sporozoites in salivary glands [11, 16, 27]. That explains why vector species remain unidentified for the great majority of avian Haemoproteus and other haemosporidian species, which is particularly true on the level of their numerous genetic lineages. If phylogenies based on cyt b gene indicate Culicoides spp. vectors, such knowledge will increase vector studies of avian haemoproteids because the methodology of generation of cyt b sequence information is relatively inexpensive, easy and well-developed in many laboratories [17–22, 53–59]. We recommend using cyt b gene phylogenies for prediction vector groups (Ceratopogonidae or Hippoboscidae) of avian haemoproteids. That will increase the determination of potential vectors. However, the application of sporozoite detection methods is crucial for final identification of vectors due to possible abortive sporogonic development, which PCR-based methods do not read [11].
Third, the available experimental data show that the same species of biting midges can transmit numerous Parahaemoproteus species, as is the case with C. impunctatus. This biting midge is a competent vector of H. belopolskyi, H. dolniki, H. parabelopolskyi, H. lanii, H. fringillae, H. balmorali, H. noctuae and H. syrnii [5,16,this study]. Additional examples are H. danilewskii, H. fringillae. H. mansoni and H. velans; these parasites complete sporogony in the biting midge Culicoides sphagnumensis [5]. Interestingly, Atkinson [27] proved experimentally the low vector specificity of Haemoproteus mansoni (probable synonym is H. meleagridis), which completes sporogony in five species of Culicoides biting midges. Lack of the strict specialisation in some Haemoproteus parasites to complete sporogony in certain species of biting midges and the broad distribution of many species of birds can explain wide geographic distribution of many lineages of avian haemoproteids [17, 18, 21, 54, 58].
It is worth mentioning that morphology of sporogonic stages of H. noctuae remains the same during development in different species of biting midges, indication that some morphological characteristics of sporogonic stages can be applied in taxonomic research on species level of haemoproteids. Ookinetes of many Parahaemoproteus species can be distinguished morphologically [5, 59]. This study shows that H. noctuae and H. syrnii can be readily distinguished due to the different lengths of their sporozoites (Table 1), providing an opportunity to distinguish these parasites at sporozoite stage in vectors. Importantly, the length of H. noctuae sporozoites was the same during development in C. impunctatus and C. nubeculosus, supporting the taxonomic value of this character. Presence of unique pedunculated oocysts in chicken malaria parasite Plasmodium juxtanucleare [60] shows that oocyst morphology is also worth attention in wildlife haemosporidian research. Additional studies are needed for determining taxonomic value of sporogonic stages, which have been insufficiently applied in classification and species identification of haemosporidian parasites.
A recent study [33], which is based on observation of single louse fly Ornithomyia sp. sampled on one tawny owl, speculated that H. syrnii might be transmitted by this hippoboscid fly. These authors observed and illustrated gametocytes and ookinetes of H. syrnii in a preparation made from the naturally infected fly, which was squashed on a glass slide. However, the observation of these parasite stages in the fly does not necessarily indicate competent vector. When blood-sucking insects take blood meal on an infected bird, mature gametocytes are ingested. The latter readily produce gametes, and ookinetes develop in non-vector blood-sucking insects and even in vitro [5, 59]. Our study shows that both H. noctuae and H. syrnii readily exflagellate and produce ookinetes in vitro (Fig. 3). Thus, it is not unexpected to observe them in engorged hippoboscids. Demonstration of sporozoites in salivary glands is an essential step for definitive demonstration that insects can act as vectors [11, 16, 24, 27, 49]. These authors [33] believed that they observed sporozoites in this fly, but provided illustration of a massive bacterial infection, which they attributed to sporozoites (Figures 32–34 in [33]). Similar bacterial infection has been seen in preparations of blood-sucking dipteran insects, where entire bodies were squashed on slides (G. Valkiūnas, personal observation). Because our experiments provided evidence about complete sporogony both of H. syrnii and H. noctuae in two species of Culicoides biting midges (Fig. 2), we question conclusions about vectorial capacity of hippoboscid flies in transmission of haemoproteids of owls. It seems unlikely that the same Haemoproteus sp. could be transmitted both by ceratopogonids and hippoboscids because transmission of the same haemosporidian species by vectors belonging to different families of dipteran insects has not been reported in haemosporidian parasites [5, 22, 52], however this hypothesis requires experimental testing [27].
Conclusions
This study adds two species of avian haemoproteids to the list of parasites transmitted by biting midges belonging to Culicoides. We show that widespread owl haemoproteids H. noctuae and H. syrnii complete sporogony in the biting midges C. impunctatus and C. nubeculosus, which are widespread in Europe and are likely to be the natural vectors of these infections. Both parasite species are characterised by rapid sporogony at relatively low temperature; that probably contributes to their spread in countries not only with warm, but also cold climates. We report that morphology of sporozoites of some Parahaemoproteus species remains the same when parasite develops into different species of biting midges, but morphology of sporozoites differs in different parasite species during their development in the same species of midges. Thus, morphology of sporozoites can be applied in some Parahaemoproteus parasite identification in vectors. This study shows that phylogenies based on cyt b gene indicate parasite-vector relationships, and we recommend using them in planning research on avian haemoproteid vectors in wildlife.
References
Miltgen F, Landau I, Ratanaworabhan N, Yenbutra S. Parahaemoproteus desseri n. sp.; Gamétogonie et shizogonie chez I’hôte naturel: Psittacula roseate de Thailande, et sporogonie experimentale chez Culicoides nubeculosus. Ann Parasitol Hum Comp. 1981;56:123–30.
Atkinson CT, Forrester DJ, Greiner EC. Pathogenicity of Haemoproteus meleagiris (Haemosporina: Haemoproteidae) in experimentally infected domestic turkeys. J Parasitol. 1988;74:228–39.
Cardona CJ, Ihejirika A, McClellan L. Haemoproteus lophortyx infection in bobwhite quail. Avian Dis. 2002;46:249–55.
Marzal A, de Lopes F, Navarro C, Moller AP. Malarial parasites decrease reproductive success: an experimental study in a passerine bird. Oecologia. 2005;142:541–5.
Valkiūnas G. Avian malaria parasites and other haemosporidia. Boca Raton: JAV; 2005.
Ferrell ST, Snowden K, Marlar AB, Garner M, Lung NP. Fatal hemoprotozoal infections in multiple avian species in a zoological park. J Zoo Wildl Med. 2007;38:309–16.
Donovan TA, Schrenzel M, Tucker TA, Pessier AP, Stalis IH. Hepatic haemorrhage, hemocoelom, and sudden death due to Haemoproteus infection in passerine birds: eleven cases. J Vet Diagn Invest. 2008;20:304–3013.
Olias P, Wegwlin M, Zenker W, Freter S, Gruber AD, Klopfleisch R. Avian malaria deaths in parrots. Eur Emerg Infect Dis. 2011;17:950–2.
Pacheco MA, Escalante AA, Garner MM, Bradley GA, Aquilar RF. Haemosporidian infection in captive masked bobwhite quail (Colinus virginianus ridgwayi), an endangered subspecies of the northen bobwhite quail. Vet Parasitol. 2011;182:113–20.
Cannell BL, Krasnec KV, Campbell K, Jones HI, Miller RD, Stephens N. The pathology and pathogenicity of a novel Haemoproteus spp. infection in wild Little Penguins (Eudyptula minor). Vet Parasitol. 2013;197:74–84.
Valkiūnas G, Palinauskas V, Ilgūnas M, Bukauskaite D, Dimitrov D, Bernotienė R, et al. Molecular characterization of five widespread avian haemosporidian parasites (Haemosporida), with perspectives on the PCR-based detection of haemosporidians in wildlife. Parasitol Res. 2014;113:2251–63.
Bennett GF, Whiteway M, Woodworth-Lynas C. A host-parasite catalogue of the avian haematozoa. Mem Univ of Nfld Occ Pap Biol. 1982;5:1–243.
Atkinson CT. Haemoproteus. In: Atkinson CT, Thomas NJ, Hunter BC, editors. Parasitic Diseases of Wild Birds. Ames: Wiley-Blackwell; 2008.
Santiago-Alarcon D, Palinauskas V, Schaefer HM. Diptera vectors of avian Haemosporidian parasites: untangling parasite life cycles and their taxonomy. Biol Rev Camb Philos Soc. 2012;87:928–64.
Valkiūnas G, Iezhova TA, Evans E, Carlson JS, Martinez-Gomez JE, Seghal RNM. Two new Haemoproteus species (Haemosporida: Haemoproteidae) from Columbiform birds. J Parasitol. 2013;99:513–21.
Žiegytė R, Palinauskas V, Bernotienė R, Iezhova TA, Valkiūnas G. Haemoproteus minutes and Haemoproteus belopolskyi (Haemoproteidae): complete sporogony in the biting midge Culicoides impunctatus (Ceratopogonidae), with implications on epidemiology of haemoproteosis. Exp Parasitol. 2014;145:74–9.
Bensch S, Hellgren O, Pérez-Tris J. A public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol Ecol Resour. 2009;9:1353–8.
Braga EM, Silveira P, Belo NO, Valkiūnas G. Recent advances in the study of avian malaria: An overview with an emphasis on the distribution of Plasmodium spp. in Brazil. Mem Inst Oswaldo Cruz. 2011; Suppl 1: 3–11.
Loiseau C, Harrigan RJ, Robert A, Bowie RC, Thomassen HA, Smith TB, et al. Host and habitat specialization of avian malaria in Africa. Mol Ecol. 2012;21:431–41.
Zehtindjiev P, Križanauskienė A, Scebba S, Dimitrov D, Valkiūnas G, Hegemann A, et al. Haemosporidian infections in skylarks (Alauda arvensis): a comparative PCR-based and microscopy study on the parasite diversity and prevalence in southern Italy and the Netherlands. Eur J Wildlife Res. 2012;58:335–44.
Clark NJ, Clegg SM, Lima MR. A review of global diversity in avian haemosporidians (Plasmodium and Haemoproteus: Haemosporida): new insights from molecular data. Int J Parasitol. 2014;44:329–38.
Perkins SL. Malaria’s many mates: past, present and future of the systematics of the order Haemosporida. J Parasitol. 2014;100:11–25.
Desser SS, Bennett GF. The genera Leucocytozoon, Haemoproteus and Hepatocystis. In: Kreier JP, Baker JR, editors. Parasitic Protozoa. 2nd ed. New York: Academic; 1993.
Valkiūnas G, Liutkevičius G, Iezhova TA. Complete development of three species of Haemoproteus (Haemosporida, Haemoproteidae) in biting midge Culicoides impunctatus (Diptera, Ceratopogonidae). J Parasitol. 2002;88:846–68.
Martinez-de la Puente J, Martinez J, Rivero-de Aguilar J, Herrero J, Merino S. On the specificity of avian blood parasites: revealing specific and generalist relationships between haemosporidians and biting midges. Mol Ecol. 2011;20:3275–87.
Levin II, Valkiūnas G, Iezhova TA, O’Brien SL, Parker PG. Novel Haemoproteus species (Haemosporida: Haemoproteidae) from the swallowtailed gull (Lariidae), with remarks on the host range of hippoboscidtransmitted avian haemoproteids. J Parasitol. 2012;98:847–54.
Atkinson CT. Vectors, epizootiology, and pathogenicity of avian species of Haemoproteus (Haemosporina: Haemoproteidae). Bull Soc Vector Ecol. 1991;16:109–26.
McClure HE, Poonswad P, Greiner EC, Laird M. Haematozoa in the birds of Eastern and Southern Asia. St. John’s: Memorial University of Newfoundland; 1978.
Valkiūnas G, Iezhova T. Parasitic Protozoa of the blood of birds in the USSR. (5. Haemoproteidae of Strigiformes and Falconiformes). Lietuvos TSR MA darbai C serija. 1989;3:49–65. in Russian.
Ishak F, Guillaumot L, Clegg SM, Phillimore AB, Black RA, Owens IPE, et al. Avian haematozoan parasites and their associations with mosquitoes across Southwest Pacific Islands. Mol Ecol. 2008;17:4545–55.
Krone O, Waldenström J, Valkiūnas G, Lessow O, Müller K, Iezhova TA, et al. Haemosporidian blood parasites in European birds of prey and owls. J Parasitol. 2008;94:709–15.
Evans M, Otter A. Fatal combined infection with Haemoproteus noctuae and Laucocytozoon ziemanni in juvenile snowy owls (Nyctea scandiaca). Vet Rec. 1998;143:72–6.
Karadjian G, Puech MP, Duval L, Chavatte JM, Snounou G, Landau I. Haemoproteus syrnii in Strix aluco from France: morphology, stages of sporogony in ahippoboscid fly, molecular characterization and discussion on the identification of Haemoproteus species. Parasite. 2013;20:32.
Carpenter S, Groschup MH, Garros C, Felippe-Bauer ML, Purse BV. Culicoides biting midges, arboviruses and public health in Europe. Antiviral Res. 2013;100:102–13.
Erik WW. The big trap for mass bird-trapping. Proc Zool Inst (St Petersburg). 1967;40:51–5 (in Russian).
Liutkevičius G. The new data on the epidemiology of bird haemoproteids (Haemosporida: Haemoproteidae) on the Curonian Spit. Acta Zool Lit. 2000;2:72–7.
Boorman J. The maintenance of laboratory colonies of Culicoides variipennis (Coq.), C. nubeculosus (Mg.) and C. riethi Kieff. (Diptera, Ceratopogonidae). B Entomol Res. 1974;64:371–7.
Gutsevich AV. The bloodsucking midges (Ceratopogonidae), in Fauna of the USSR. Dipteran insects, vol. 3, part 5. Leningrad: Nauka Press; 1973 (in Russian).
Richardson DS, Jury FL, Blaakmeer K, Komdeur J, Burke T. Parentage assignment and extra-group paternity in a cooperative breeder: the Seychelles warbler (Acrocephalus sechellensis). Mol Ecol. 2001;10:2263–73.
Bensch S, Stjernman M, Hasselquist D, Ostman O, Hansson B, Westerdahl H, et al. Host specifity in avian blood parasites: a study of Plasmodium and Haemoproteus mitochondrial DNA amplified from birds. Proc R Soc B. 2000;267:1583–9.
Hellgren O, Waldenström J, Bensch S. A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium and Haemoproteus from avian blood. J Parasitol. 2004;90:797–802.
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome C oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–9.
Hall TA. A user-friendly biological sequence alignment editor and analysis program for Windows 98/98/NT. Nucleic Acid Symp Ser. 1999;41:95–8.
Ronquist F, Heulsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4.
Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–8.
Tamura K, Dubley J, Nei M, Kumar S. Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9.
Fallis AM, Bennett GF. Ceratopogonidae as intermediate hosts for Haemoproteus and other parasites. Mosq News. 1961;21:21–8.
Fallis AM, Bennett GF. Sporogony of Leucocytozoon and Haemoproteus in simuliids and ceratopogonids and a revised classification of the Haemosporidiida. Can J Zool. 1961;39:215–28.
Fallis AM, Wood DM. Biting midges (Diptera: Ceratopogonidae) as intermediate hosts for Haemoproteus of ducks. Can J Zool. 1957;35:425–35.
Khan RA, Fallis AM. A note on the sporogony of Parahaemoproteus velans (Haemoproteus velans Coatney and Roudabush) (Haemosporidia: Haemoproteidae) in species of Culicoides. Can J Zool. 1971;49:420–1.
Levin II, Valkiūnas G, Santiago-Alarcon D, Cruz LL, Iezhova TA, O’Brien SL, et al. Hippoboscid-transmitted Haemoproteus parasites (Haemosporida) infect Galapagos pelecaniform birds: evidence from molecular and morphological studies, with a description of Haemoproteus iwa. Int J Parasitol. 2011;15:1019–27.
Garnham PCC. Malaria parasites and other haemosporidia. Oxford: Blackwell; 1966.
Martinsen ES, Perkins SL, Schall JJ. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): evolution of life-history traits and host switches. Mol Phylogenet Evol. 2008;47:261–73.
Marzal A, Ricklefs RE, Valkiūnas G, Albayrak T, Arriero E, Bonneaud C, et al. Diversity, loss and gain of malariae parasites in a globally invasive bird. PLoS One. 2011;6:e21905.
Matta NE, Pacheco MA, Escalante AA, Valkiūnas G, Ayerbe-Quiñones F, Acevedo-Cendales LD. Description and molecular characterization of Haemoproteus macrovacuolatus n. sp. (Haemosporida, Haemoproteidae), a morphologically unique blood parasite of black-bellied whistling duck (Dendrocygna autumnalis) from South America. Parasitol Res. 2014;113:2991–3000.
Walther EL, Valkiūnas G, González AD, Matta NE, Ricklefs RE, Cornel A, et al. Description, molecular characterization, and patterns of distribution of a widespread New World avian malaria parasite (Haemosporida: Plasmodiidae), Plasmodium (Novyella) homopolare sp. nov. Parasitol Res. 2014;113:3319–32.
Dimitrov D, Palinauskas V, Iezhova TA, Bernotienė R, Ilgūnas M, Bukauskaitė D, et al. Plasmodium spp.: an experimental study on vertebrate host susceptibility to avian malaria. Exp Parasitol. 2015;148:1–16.
Pérez-Rodríguez A, de la Puente J, Onrubia A, Pérez-Tris J. Molecular characterization of haemosporidian parasites from kites of the genus Milvus (Aves: Accipitridae). Int J Parasitol. 2013;43:381–7.
Dimitrov D, Valkiūnas G, Zehtindjiev P, Ilieva M, Bensch S. Molecular characterization of haemosporidian parasites (Haemosporida) in yellow wagtail (Motacilla flava), with description of in vitro ookinetes of Haemoproteus motacillae. Zootaxa. 2013;3666:369–81.
Bennett GF, Warren M, Cheong WH. Biology of the Malaysian strain of Plasmodium juxtanucleare Versiani and Gomes, 1941. II. The sporogonic stages in Culex (Culex) sitiens Wiedmann. J Parasitol. 1966;52:647–52.
Acknowledgements
We thank the staff of the Institute for Animal Health, Pirbright Laboratory, UK for providing C. nubeculosus midges for establishing their colony, director of the Biological Station “Rybachy” Casimir V. Bolshakov, for generously providing facilities for the experimental research, and the staff of the Station, for assistance in the field. This research was supported by the Open Access to research infrastructure of the Nature Research Centre under Lithuanian open access network initiative. Research Council of Lithuania has funded publication of this article.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Experimental conception and design: GV, DB, RŽ; biting midge fieldwork: DB, RŽ, VP, DD, MI, RB, GV; biting midge dissection and laboratory maintenance: DB, RŽ, RB; donor bird collection and testing: MYM, TAI, GV; phylogenetic analysis and its discussion VP, DB, GV; paper writing: DB, GV. All the authors read and approved the final version of the manuscript.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Bukauskaitė, D., Žiegytė, R., Palinauskas, V. et al. Biting midges (Culicoides, Diptera) transmit Haemoproteus parasites of owls: evidence from sporogony and molecular phylogeny. Parasites Vectors 8, 303 (2015). https://doi.org/10.1186/s13071-015-0910-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-015-0910-6