Abstract
Background
Habitat types can affect vector and pathogen distribution and transmission dynamics. The prevalence and genetic diversity of Plasmodium spp. in two eastern chimpanzee populations—Kalinzu Forest Reserve, Uganda and Issa Valley, Tanzania—inhabiting different habitat types was investigated. As a follow up study the effect of host sex and age on infections patterns in Kalinzu Forest Reserve chimpanzees was determined.
Methods
Molecular methods were employed to detect Plasmodium DNA from faecal samples collected from savanna-woodland (Issa Valley) and forest (Kalinzu Forest Reserve) chimpanzee populations.
Results
Based on a Cytochrome-b PCR assay, 32 out of 160 Kalinzu chimpanzee faecal samples were positive for Plasmodium DNA, whilst no positive sample was detected in 171 Issa Valley chimpanzee faecal samples. Sequence analysis revealed that previously known Laverania species (Plasmodium reichenowi, Plasmodium billbrayi and Plasmodium billcollinsi) are circulating in the Kalinzu chimpanzees. A significantly higher proportion of young individuals were tested positive for infections, and switching of Plasmodium spp. was reported in one individual. Amongst the positive individuals sampled more than once, the success of amplification of Plasmodium DNA from faeces varied over sampling time.
Conclusion
The study showed marked differences in the prevalence of malaria parasites among free ranging chimpanzee populations living in different habitats. In addition, a clear pattern of Plasmodium infections with respect to host age was found. The results presented in this study contribute to understanding the ecological aspects underlying the malaria infections in the wild. Nevertheless, integrative long-term studies on vector abundance, Plasmodium diversity during different seasons between sites would provide more insight on the occurrence, distribution and ecology of these pathogens.
Similar content being viewed by others
Background
Parasite distribution and transmission dynamics are influenced by the ecological context of the host-parasite interactions and a variety of local environmental parameters [1–3]. In the case of vector-borne Plasmodium infections, the primary effect of habitat on the transmission of malaria is by affecting larvae development, abundance and distribution of competent vectors [4–7]. Numerous studies have demonstrated the relationship between specific habitats and levels of Plasmodium infections in humans [8–12]. However, research addressing habitat types as a source of variation in prevalence and diversity of these parasites in wild apes is lacking. In addition to habitat, host traits such as age, sex and host density may also have an influence on host parasite infection and transmission of Plasmodium spp. [13–15].
Chimpanzees (Pan troglodytes), like several other primates, harbour a multitude of malaria parasites. With the development and refinement of molecular diagnostic techniques together with non-invasive sampling, at least seven distinct Plasmodium species are known to infect wild chimpanzees. Four of them, Plasmodium reichenowi, Plasmodium gaboni, Plasmodium billcollinsi and Plasmodium billbrayi belong to the subgenus Laverania and are chimpanzee-host specific [16–22]. The remaining three species, usually referred to as Plasmodium malariae-like, Plasmodium ovale-like and Plasmodium vivax-like, rarely occur in chimpanzees and they are genetically related to their human counterparts. Nevertheless, the nomenclature of these rare taxa requires further investigation [19]. Given the high genetic diversity of Plasmodium species reported from chimpanzees and other primates including humans [19, 20, 23], a better understanding of the infection dynamics and interactions between parasites, Anopheles mosquitoes, hosts and environmental parameters that facilitate malaria transmission in apes is required [15, 18, 24].
In the current study, the prevalence and genetic diversity of Plasmodium spp. was investigated in two populations of eastern chimpanzees (P. t. schweinfurthii) inhabiting two different habitats: (1) savanna woodlands in Issa Valley, Tanzania and (2) evergreen moist forest in Kalinzu Forest Reserve (KFR), Uganda. Malaria infection was compared between these two habitats (savanna and moist evergreen forest) because of their variable environmental parameters that may influence the exposure to malaria parasites with varying degrees in chimpanzee populations. Because chimpanzees at KFR are habituated, the relationship between age, sex and malaria infection patterns in this population was additionally addressed.
Methods
Study sites
Issa Valley, Tanzania
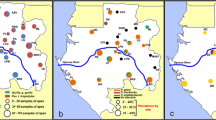
The Issa valley is located in western Tanzania (Fig. 1), about 90 km east of Mahale Mountains National Park, and approximately 70 km from Uvinza, the nearest legitimate village. Issa valley is characterised as an open area with no formal protective status, where small-scale illegal human activity for hunting and logging takes place [25]. The entire region is one of the driest and most open chimpanzee habitats, with an altitudinal range of 900–1800 m above sea level [26]. There is an extended dry season (May–September), with rains from October–April, peaking in January (unpublished data), averaging 1095 mm/year (range 835–1395 mm/year). Average daily temperature varies from 11–35 °C [27]. The habitat is dominated by savanna (Miombo) woodland, characterized by Brachystegia and Julbernardia trees, with small riparian forest patches [26]. The population density of Issa chimpanzees is estimated to be ~0.25 individuals/km2 [25]. Data on the prevalence of P. vivax in this population have been reported elsewhere [28]. In addition to chimpanzees, several other primate species inhabit the study site, including red colobus monkeys (Piliocolobus tephrosceles), yellow baboons (Papio cynocephalus), blue (Cercopithecus mitis) and red-tailed monkeys (Cercopithecus ascanius), vervet monkeys (Chlorocebus pygerythrus), bushbabies (Galago senegalensis, Cercopithecus moholi) and greater galagos (Otolemur crassicaudatus) [26].
KFR, Uganda
Kalinzu is one of the three largest forest blocks in Uganda. The forest reserve (~137 km2) is located on the eastern ridge of the western Rift valley in western Uganda (Fig. 2), with an altitudinal range of 1200–1500 m above sea level [29]. The area is adjacent to Kashoha–Kitomi Forest Reserve and Maramagambo Forest Reserve on the north and west sides, agricultural fields to the east and tea plantations to the south [29]. Kalinzu has a bimodal distribution of rainfall with peaks between September–December and March–May, and average annual rainfall of 1584 mm. The average daily temperature varies from 15 to 25 °C [30, 31]. The vegetation is classified as medium altitude moist evergreen forest, with common species including Musangaleo and Ficus spp. [32]. The chimpanzee population density is estimated to be ~1.67 individuals/km2 [33]. In addition to Pan troglodytes schweinfurthii, black and white colobus (Colobus guereza), olive baboons (Papio anubis), red tailed (Cercopithecus ascanius), blue (C. mitis), and L’hoests monkeys (C. lhoesti) occur in the area [32].
Sample collection
Issa Valley
171 faecal samples were collected from a single community of chimpanzees inhabiting the Issa study area between March–May 2012 and June–August 2013. It was not possible to attribute the faecal samples to specific individuals. Most of the faecal samples were collected underneath fresh nests (~12 h old) and some from chimpanzee trails. Approximately 20 g of faecal material was collected in a 50 ml tube, containing 20 ml of RNAlater™ (Ambion Inc., Austin, TX). All faecal samples were stored in a freezer at −20 °C on site, and subsequently shipped to the Czech Republic, where they were kept at −20/−80 °C until DNA extraction.
KFR
Between April and July 2014, faecal samples collected from 41 habituated chimpanzees (males, n = 20; females, n = 21). A total of 123 fresh faecal samples, ranging from 1 to 10 faecal samples per individual were collected. Samples were collected during direct observations of chimpanzees. Concurrently, during tracking of chimpanzees 37 faecal samples were collected from unidentified individuals. Collection and storage protocols were the same as those at Issa, with the exception that samples were kept at 4 °C in a fridge at base camp prior to shipping to the Czech Republic, where they were kept at −20/−80 °C until DNA extraction.
Molecular methods
Total DNA was extracted from 1.5 ml of the faecal—RNAlater™ suspension using a QIAamp Stool DNA Mini kit (Qiagen, Valencia, CA, USA) and PSP® Spin Stool DNA Kit (Stratec Molecular, Germany) according to the manufacturer’s protocol. Bound DNA was eluted in 100 μl elution buffer. To determine the concentration of the extracted DNA, total DNA was measured by fluorometry, using a Qubit (Invitrogen, Carlsbad, CA, USA). To screen samples for Plasmodium, a nested PCR was performed on each sample targeting a ~930 bp fragment of the Plasmodium cytochrome b (cyt-b) gene, as described by Prugnolle et al. [34], with modification of the second PCR reaction. A pair of short internal primers amplifying overlapping fragments (516 and 558 bp) was designed, retrieved sequences were contiged to obtain same region of cyt-b. First round PCRs were performed in a 25 μl reaction, containing 12.5 μl of PCR mix (Qiagen), 2.5 μl of solution Q (Qiagen) and 0.2 μl of each primer (DW2 and DW4) in 10 pmol concentration and 4 μl of the DNA sample. Second nested PCR was performed using two different set of reactions, using Cytb1 (5′-CTCTATTAATTTAGTTAAAGCACA-3′) and Cytb2B (5′-GCTCTATCATACCCTAAAGG-3′) in the first set, and Cytb2 (5′-ACAGAATAATCTCTAGCACC-3′) and Cytb1A (5′-CAAATGAGTTATTGGGGTGCAACT-3′) for the second set. Two µl of first round PCR product was then used in a second round 25 µl nested PCR reaction, containing 12.5 µl common Master Mix (Top-Bio, Czech Republic) and 1 µl of each primer in 10 pmol concentration. For details of the modified nested PCR conditions see [15]. PCR products were visualized on 2 % agarose gel and stained with Gold-View. Bands of the expected size were visualized using an UV light source, excised, purified using QIAquick gel extraction kit (Qiagen, Germany) and sequenced in both directions using internal primers by Macrogen capillary sequencing services (Macrogen Europe, the Netherlands).
Sequence and phylogenetic analyses
Sequences were edited in Chromas Pro 1.5 software (Technelysium, Ltd) and alignment was prepared with ClustalW multiple alignment tool implemented in Bioedit Sequence Alignment Editor v.7.0.9.1 [35]. All suitable retrieved sequences were submitted to GenBank™ database under the Accession Numbers KT864824–KT864842.
The alignment was checked manually and the resulting sequence were (~758 bp) later used for phylogenetic analyses. To examine the phylogenetic relationship of the new dataset, sequences from different ape Plasmodium species downloaded from GenBank™ were added to the final alignment. For the final analyses, only haplotypes were further included (haplotypes and redundant sequences are shown in Table 1).
Phylogenetic relationships were inferred using the maximum likelihood (ML) method under the general time-reversible evolutionary model with gamma distributed substitution rates (GTR + Γ) in program PhyML 3.0 [36]. Nodal support was assessed by bootstrap using 1000 pseudoreplicates. Additionally, Bayesian methods using the program MrBayes 3.2.2 [37] was also used to reconstruct phylogenetic relationships. Setting for the evolutionary model was the same as in ML and the search was carried out in two simultaneous runs of one million generations, sampled each 100 generations, with a burn-in of 25 %.
Cloning of mixed infection samples
Two samples were cloned separately with a TOPO® TA cloning kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Plasmids containing inserts were isolated from positive Escherichia coli colonies by GenElute™ plasmid mini prep kit (Sigma-Aldrich, St. Louis, MO, USA). DNA extracts from at least six randomly selected colonies were sequenced in both directions.
Statistical analyses
Prevalence was defined as the number of Plasmodium-positive individuals divided by the total of individuals tested. Samples collected from unidentified individuals were not included for the calculation of prevalence, but they were used to investigate the genetic diversity of the parasites. Of the 41 habituated individuals sampled in KFR, 25 were re-sampled to observe the fluctuation of the infections. In order to examine the possible effect of sex and age on the occurrence of malaria in KFR chimpanzees, a general linear mixed model (GLMM) with binomial distribution was fitted. Since there were a limited number of faecal samples from juveniles and subadults, age classes were pooled and grouped as juveniles/subadults and adults. Age-classes were verified based on previously suggested categorization [38]. Samples were classified according to sex (fixed factor: male, female) and class of age (fixed factor: juvenile/subadult, adult). Individual identity was treated as a random factor. Statistical analyses were performed in R [39].
Results
In total, 331 chimpanzee faecal samples (Table 2) from Issa Valley and KFR were examined. All faecal samples collected from Issa chimpanzees were negative for Plasmodium DNA. On the contrary, Plasmodium spp. was detected in 32 out of 160 (both identified and unidentified individuals) faecal samples collected from KFR chimpanzees. In total, 22 out of 123 samples collected from identified individuals were positive for Plasmodium DNA;10 out of 37 samples from unidentified individuals were Plasmodium-positive. The prevalence among identified individuals was 43.9 % (n = 18/41). The general linear mixed model showed that sex had no significant effect on the susceptibility to infection (GLMM: z = −0.027, p = 0.283), while age was a significant factor influencing Plasmodium infection. The total prevalence of Plasmodium spp. was significantly higher among juvenile/subadult individuals than adults (GLMM: z = 2.308, p = 0.020). Of the re-sampled individuals (n = 25), eleven were found positive at least once. Variation on detection of Plasmodium DNA (negative-to-positive and vice versa) was common and observed in 18 identified individuals (Table 3). Switching of Plasmodium spp. was observed in one individual (Table 3).
Alignment and phylogenetic analysis of the obtained cyt-b sequences (both from identified and unidentified individuals) with reference sequences indicated the presence of Plasmodium strains that specifically infect only chimpanzees (see Additional file 1). Among the retrieved sequences, 12 were P. reichenowi, 11 P. billbrayi and seven P. billcollinsi. All sequences obtained in this study clustered with their homologous sequences retrieved from GenBank™ and form well-supported clades. Geographical sub-structuring among P. reichenowi was observed, whereby sequences obtained from P. t. schweinfurthii clustered separately from other P. reichenowi sequences from P. t. troglodytes and Pan troglodytes ellioti. No samples containing cyt-b of P. gaboni or non-Laverania species (P. vivax-like, P. malariae-like and P. ovale-like) were detected in this dataset. Mixed infections were detected in two samples. Sequences of two PCR amplicons showed double peaks in the chromatograms, suggesting mixed infections.
These samples were further processed by cloning to identify Plasmodium to species level. In the first sample (from an unidentified individual), 15 sequences were obtained with two representative sequence patterns that were in agreement with BLAST-searches for the cyt-b sequences: 14 sequences were 99–100 % similar to P. reichenowi (acc. number: HM235389), and one sequence was 99 % similar to P. billbrayi (acc. GQ355468). In the second sample (from an identified individual), 12 sequences were obtained with three representative sequences patterns: four sequences were 99 % similar to P. reichenowi (acc. number: HM235389), five sequences were 99–100 % to P. billbrayi (acc. number: GQ355468), and three sequences were 99 % similar to P. billcollinsi (acc. number: HM235392).
Discussion
A number of studies have described the distribution and genetic diversity of Plasmodium spp. in African great apes [17, 18, 22, 34, 40, 41], yet there is substantial lack of knowledge on the effect of intrinsic and extrinsic factors that govern malaria parasite transmission and frequencies of infections in free ranging chimpanzees. This is the first study to investigate the prevalence and genetic diversity of Plasmodium spp. in KFR. The findings from KFR are comparable to previous studies by Liu et al. [18], that were conducted at multiple field sites, as well as to the study by Kaiser et al. [41] from Budongo Forest in Uganda. While no Plasmodium spp was detected from Issa Valley samples, results from a previous study [28] revealed that four out of three hundred thirteen chimpanzee samples from this population to be positive for P. vivax-like. Variation in the prevalence between this study and that of Liu et al. [28] is most likely to be attributable to smaller sample set, and, possibly also to differences in sensitivity of detection methods. Looking at this discrepancy from a different perspective, P. vivax tends to stay dormant in the liver for many years [42]. Consequently, it can be speculated that during sampling time shedding of Plasmodium DNA into the intestinal lumen was minimal, leading to failure to detect P. vivax DNA in faecal samples.
An overall prevalence of Plasmodium spp. in KFR was 43.9 %, while all faecal samples from Issa Valley were negative. The remarkable ecological differences between KFR and Issa Valley habitats represent most plausible explanation for observed differences, as they may impact on the species diversity and abundance of anopheline mosquitoes. However, also host density may have significant impact on the transmission and maintenance of infections in a given population [12]. Kalinzu chimpanzees live at a relatively high density (~1.67 individuals/km2, [33]) compared to Issa chimpanzees (~0.25 individuals/km2, [25]). Then, the abundance of hosts may act as an additional factor influencing the prevalence of Plasmodium spp.
Liu et al. [28] screened another but forest-inhabiting eastern chimpanzee population (Pan t. schweinfurthii) from Gombe National Park, and none of the samples was positive for P. vivax-like. The absence (or very low prevalence Liu et al. [28]) of Plasmodium infection is these eastern chimpanzee populations (Issa Valley and Gombe National Park) could be also attributed to the genetic factors related to hosts as observed in human [43] rather than to their habitat. Unfortunately, it is difficult to reliably compare the results of these two studies due to the different diagnostic techniques employed (P. vivax species-specific assay in the Gombe study [28], and Plasmodium genus-specific in the present study). Nevertheless, screening of near-by forested (Mahale Mountains National Park) and other savanna-dwelling chimpanzees (e.g. Semliki, Uganda; Fongoli, Senegal), as well as re-screening of the Gombe chimpanzee population for presence of Laverania species would offer an insight into the factors the influence the occurrence of Plasmodium spp. in eastern chimpanzees.
Over the past 5 years, numerous Plasmodium species have been reported to circulate in free-ranging great apes [19]. Consistent with previous studies [18, 22, 34, 41], sequence analyses of the cyt-b gene of Plasmodium spp. from Kalinzu chimpanzees revealed a high diversity of malaria parasites. With the exception of P. gaboni, which was not detected in this sample set, most of the sequences were identified as P. billbrayi, however, P. reichenowi and P. billcollinsi were also confirmed. Phylogenetic analysis showed that all sequences in the present study cluster within the clades of subgenus Laverania, no sequence belonging to non-Laverania (P. vivax-like, P. ovale-like and P. malariae-like) lineage was identified. These results agree with recent findings on ape malaria, where Laverania lineages were the only ones reported from central chimpanzees across multiple field sites in Gabon [22], although, non-Laverania parasites are known to circulate within the same chimpanzee populations [44].
In initial phylogenetic analysis, a geographical sub-structuring in P. reichenowi related to host phylogeography appeared (Fig. 3). A phylogram resulting from the extended dataset confirmed this sub-structure. All P. reichenowi sequences obtained from P. t. schweinfurthii formed a separated subclade as previously observed by Liu et al. [18]. This sub-structuring could be influenced by the geographical barriers or differences in mosquito vectors responsible for transmission of malaria parasites. Further investigation into ape-malaria from other chimpanzee populations, as well as the inclusion of environmental factors that may influence Plasmodium species distribution and abundance in wild great apes, will further contribute to a better understanding of Plasmodium species diversity and dynamics.
Of the two host traits analysed in this study, only age was found to be statistically significant, with young chimpanzees more likely to be infected with Plasmodium spp. than older ones. A similar trend was observed in western chimpanzees of Taï, Ivory Coast [14], western lowland gorillas inhabiting Dzanga-Sangha Protected Areas [15], as well as in humans [45, 46]. The time needed to develop semi-immunity against the malaria parasite may explain why Plasmodium was encountered more frequently among younger individuals [47]. Also the failure to find differences in infection levels between the sexes is consistent with previous results from western lowland gorillas [15] and western chimpanzees [14]. Indeed, the scarcity of information about the biology and ecology of Laverania lineages and their interactions with hosts, preclude us from drawing a precise picture of the infection dynamics.
The pattern of infections (negative-to-positive and vice versa) was observed in 18 individuals sampled more than once over the course of the sampling period. It is worth noting that negative samples observed in this study do not necessary reflect the absence of infections. Rather, this phenomenon might be explained by fluctuation of parasitaemia level and shedding of parasite DNA in faeces, combined with sensitivity of the Plasmodium detection in faecal samples expected to be lower compared to blood samples [18, 48]. These findings may indicate that detection of Plasmodium DNA in faeces is prone to high risk of false negativity, hindering adequate assessment of actual prevalence of malaria in free ranging chimpanzee populations.
Conclusion
The findings of this study contribute to a broader understanding of malaria occurrence among wild chimpanzees. The differences observed may result from local variation in host exposure to mosquito vectors, extrinsic factors, differences in chimpanzee density, as well as host genetic related factors. Future research should focus not only on screening chimpanzees that live in a variety of habitats, but also identifying potential vectors and vector abundance, in order to provide insights on the distribution and occurrence of Plasmodium spp. in chimpanzees.
References
Craig MH, Snow RW, le Sueur D. A climate-based distribution model of malaria transmission in sub-Saharan Africa. Parasitol Today. 1999;15:105–11.
Grover-Kopec E, Kawamo M, Klaver RW, Blumenthal B, Ceccato P, Connor SJ. An online operational rainfall-monitoring resource for epidemic malaria early warning systems in Africa. Malar J. 2005;4:6.
Blanford JI, Blanford S, Crane RG, Mann ME, Paaijmans KP, Schreiber KV, et al. Implications of temperature variation for malaria parasite development across Africa. Sci Rep. 2013;3:1300.
Kovats RS, Campbell-Lendrum DH, McMichael AJ, Woodward A, Cox JS. Early effects of climate change: do they include vector-borne disease? Philos Trans R Soc Lond B Biol Sci. 2001;356:1057–68.
Hunter PR. Climate change and waterborne and vector-borne disease. J Appl Microbiol. 2003;94:37S–46S.
McMichael AJ, Campbell-Lendrum DH, Corvalán CF, Ebi KL, Githeko AK, Scheraga JD, et al. Climate change and human health: risks and responses. Geneva: World Health Organization; 2003.
Patz JA, Campbell-Lendrum D, Holloway T, Foley JA. Impact of regional climate change on human health. Nature. 2005;438:310–7.
Dery DB, Brown C, Asante KP, Adams M, Dosoo D, Amenga-Etego S, et al. Patterns and seasonality of malaria transmission in the forest-savannah transitional zones of Ghana. Malar J. 2010;9:314.
Abonuusum A, Owusu-Daako K, Tannich E, May J, Garms R, Kruppa T. Malaria transmission in two rural communities in the forest zone of Ghana. Parasitol Res. 2011;108:1465–71.
Badu K, Brenya RC, Timmann C, Garms R, Kruppa TF. Malaria transmission intensity and dynamics of clinical malaria incidence in a mountainous forest region of Ghana. MalariaWorld J. 2013;4:14.
Songue E, Tagne C, Mbouyap P, Essomba P, Moyou-Somo R. Epidemiology of malaria in three geo-ecological zones along the Chad-Cameroon pipeline. Am J Epidemiol Infect Dis. 2013;1:27–33.
Kar NP, Kumar A, Singh OP, Carlton JM, Nanda N. A review of malaria transmission dynamics in forest ecosystems. Parasit Vectors. 2014;7:265.
Doolan DL, Dobano C, Baird JK. Acquired immunity to malaria. Clin Microbiol Rev. 2009;22:13–36.
De Nys HM, Calvignac-Spencer S, Thiesen U, Boesch C, Wittig RM, Mundry R, et al. Age-related effects on malaria parasite infection in wild chimpanzees. Biol Lett. 2013;9:20121160.
Mapua MI, Qablan MA, Pomajbíková K, Petrželková KJ, Huzová Z, Rádrová J, et al. Ecology of malaria infections in western lowland gorillas inhabiting Dzanga Sangha Protected Areas. Central Afr Repub Parasitol. 2015;142:890–900.
Ollomo B, Durand P, Prugnolle F, DouzeryE Arnathau C, Nkoghe D, et al. A new malaria agent in African hominids. PLoS Pathog. 2009;5:e1000446.
Krief S, Escalante AA, Pacheco MA, Mugisha L, André C, Halbwax M, et al. On the diversity of malaria parasites in African apes and the origin of Plasmodium falciparum from Bonobos. PLoS Pathog. 2010;6:e1000765.
Liu W, Li Y, Learn GH, Rudicell RS, Robertson JD, Keele BF, et al. Origin of the human parasite Plasmodium falciparum in gorillas. Nature. 2010;467:420–5.
Rayner JC, Liu W, Peeters M, Sharp PM, Hahn BH. A plethora of Plasmodium species in wild apes: a source of human infection? Trends Parasitol. 2011;27:222–9.
Duval L, Ariey F. Ape Plasmodium parasites as a source of human outbreaks. Clin Microbiol Infect. 2012;18:528–32.
Pacheco MA, Cranfield M, Cameron K, Escalante AA. Malarial parasite diversity in chimpanzees: the value of comparative approaches to ascertain the evolution of Plasmodium falciparum antigens. Malar J. 2013;12:328.
Boundenga L, Ollomo B, Rougeron V, Mouele LY, Mve-Ondo B, Delicat-Loembet LM, et al. Diversity of malaria parasites in great apes in Gabon. Malar J. 2015;14:111.
Singh B, Daneshvar C. Human infections and detection of Plasmodium knowlesi. Clin Microbiol Rev. 2013;26:165–84.
Verhulst NO, Smallegange RC, Takken W. Mosquitoes as potential bridge vectors of malaria parasites from non-human primates to humans. Front Physiol. 2012;3:197.
Piel AK, Lenoel A, Johnson C, Stewart FA. Deterring poaching in western Tanzania: the presence of wildlife researchers. Glob Ecol Conserv. 2015;3:188–99.
Hernandez-Aguilar RA. Chimpanzee nest distribution and site reuse in a dry habitat: implications for early hominid ranging. J Hum Evol. 2009;57:350–64.
Stewart FA. The evolution of shelter: Ecology and ethology of chimpanzee nest building. PhD thesis. Cambridge: University of Cambridge; 2011.
Liu W, Li Y, Shaw KS, Learn GH, Plenderleith LJ, Malenke JA, et al. African origin of the malaria parasite Plasmodium vivax. Nat Commun. 2014;5:3346.
Yasuoka H, Kimura D, Hashimoto C, Furuichi T. Quantitative assessment of livelihoods around great ape reserves: cases in luo scientific reserve, Drcongo, and Kalinzu Forest Reserve, Uganda. Afr Study Monogr. 2012;43:137–59.
Hashimoto C. Population census of the chimpanzees in the Kalinzu Forest, Uganda: comparison between methods with nest counts. Primates. 1995;36:477–88.
Furuichi T, Hashimoto C, Tashiro Y. Extended application of a marked-nest census method to examine seasonal changes in habitat use by chimpanzees. Inter J Primatol. 2001;22:913–28.
Howard PC. Nature Conservation in Uganda’s Tropical Forest Reserves. Gland: IUCN; 1991.
Plumptre AJ, Cox D. Counting primates for conservation: primate surveys in Uganda. Primates. 2006;47:65–73.
Prugnolle F, Durand P, Neel C, Ollomo B, Ayala FJ, Arnathau C, et al. African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum. Proc Natl Acad Sci USA. 2010;107:1458–63.
Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–8.
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21.
Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetics inference under mixed models. Bionformatics. 2003;19:1572–4.
Nishida T, Corp N, Hamai M, Hasegawa T, Hiraiwa-Hasegawa M, Hosaka K, et al. Demography, female life history, and reproductive profiles among the chimpanzees of Mahale. Am J Primatol. 2003;59:99–121.
R Development Core Team. R: a language and environment for statistical computing. Vienna: the R Foundation for Statistical Computing; 2011. ISBN 3-900051-07-0.
Duval L, Fourment M, Nerrienet E, Rousset D, Sadeuh SA, Goodman SM, et al. African apes as reservoirs of Plasmodium falciparum and the origin and diversification of the Laverania subgenus. Proc Natl Acad Sci USA. 2010;107:10561–6.
Kaiser M, Löwa A, Ulrich M, Ellerbrok H, Goffe AS, Blasse A, et al. Wild chimpanzees infected with 5 Plasmodium species. Emerg Infect Dis. 2010;16:1956–9.
Galinski MR, Meyer EV, Barnwell JW. Plasmodium vivax: modern strategies to study a persistent parasite’s life cycle. Adv Parasitol. 2013;81:1–26.
Kwiatkowski DP. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 2005;77:171–92.
Prugnolle F, Rougeron V, Becquart P, Berry A, Makanga B, Rahola N, et al. Diversity, host switching and evolution of Plasmodium vivax infecting African great apes. Proc Natl Acad Sci USA. 2013;110:8123–38.
Doolan DL, Dobano C, Baird JK. Acquired immunity to malaria. Clin Microbiol Rev. 2009;22:13–36.
De Beaudrap P, Nabasumba C, Grandesso F, Turyakira E, Schramm B, Boum Y, et al. Heterogeneous decrease in malaria prevalence in children over a six-year period in south-western Uganda. Malar J. 2011;10:132.
Pouniotis DS, Proudfoot O, Minigo G, Hanley JL, Plebanski M. Malaria parasite interactions with the human host. J Postgrad Med. 2004;50:30–4.
Nwakanma DC, Gomez-Escobar N, Walther M, Crozier S, Dubovsky F, Malkin E, et al. Quantitative detection of Plasmodium falciparum DNA in saliva, blood, and urine. J Infect Dis. 2009;199:1567–74.
Sundararaman SA, Liu W, Keele BF, Learn GH, Bittinger K, Mouacha F, et al. Plasmodium falciparum-like parasites infecting wild apes in southern Cameroon do not represent a recurrent source of human malaria. Proc Natl Acad Sci USA. 2013;110:7020–5.
Joy DA, Feng X, Mu J, Furuya T, Chotivanich K, Krettli AU, et al. Early origin and recent Expansion of Plasmodium falciparum. Science. 2003;300:318–21.
Miao M, Yang Z, Patch H, Huang Y, Escalante AA, Cui L. Plasmodium vivax populations revisited: mitochondrial genomes of temperate strains in Asia suggest ancient population expansion. BMC Evol Biol. 2012;12:22.
Hayakawa T, Arisue N, Udono T, Hirai H, Sattabongkot J, Toyama T, et al. Identification of Plasmodium malariae, a human malaria parasite, in imported chimpanzees. PLoS ONE. 2009;4:e7412.
Authors’ contributions
MIM, JB, FAS and AP collected faecal samples in the field. MIM, ES, KB, KH performed the molecular work. PV performed phylogenetic analyses. JB performed statistical analyses. HPF and MAQ supervised the laboratory work. KJP, CH and DM coordinated and designed the research project. MIM compiled the results and wrote the manuscript. KJP, DM, AKP, FAS, KH, PV, JB, HPF and MAQ edited the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We are grateful to the Tanzania Wildlife Research Institute (TAWIRI), the Tanzania Commission for Science and Technology (COSTECH), Uganda National Council for Science and Technology, Uganda Wildlife Authority and Uganda National Forest Authority for approval to conduct the field work and faecal sample collections. We would like to express our thanks to Barbora Kalousová for sample collection at Issa Valley, as well as to field assistants from both field sites. Further, we are thankful to Dr. Petr Lany from VFU for access to the P3 laboratory and Barbara Eigner and Walpurga Wille-Piazzai from the Institute of Parasitology, Department of Pathobiology, University of Veterinary Medicine Vienna, Austria for assist with laboratory work. We would like to thank also Omar Radaideh from Department of Geological Science, Masaryk University for formatting the maps.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The dataset of this article is available upon request.
Funding
This work was funded by IGA 90/2014/FVL, grant of the Czech Academy of Sciences M200961204 and the EurNegVec COST Action TD1303. This research was carried out under the project CEITEC 2020 (LQ1601) with financial support from the Ministry of Education, Youth and Sports of the Czech Republic under the National Sustainability Programme II and further co-financed from European Social Fund and state budget of the Czech Republic (project OPVK CZ.1.07/2.3.00/20.0300). The study was further supported by the Institute of Vertebrate Biology Academy of Science of the Czech Republic (RVO: 68081766) and by project LO1218 under the NPU I program. We also acknowledge a grant for the development of research organization (RVO: RO0516). Research at Issa Valley (and for the Ugalla Primate Project) is supported by the UCSD/Salk Center for Academic Research and Training in Anthropogeny (CARTA).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Mapua, M.I., Petrželková, K.J., Burgunder, J. et al. A comparative molecular survey of malaria prevalence among Eastern chimpanzee populations in Issa Valley (Tanzania) and Kalinzu (Uganda). Malar J 15, 423 (2016). https://doi.org/10.1186/s12936-016-1476-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-016-1476-2