Abstract
Background
Malaria is causing more than half of a million deaths and 214 million clinical cases annually. Despite tremendous efforts for the control of malaria, the global morbidity and mortality have not been significantly changed in the last 50 years. Artemisinin, extracted from the medicinal plant Artemisia sp. is an effective anti-malarial drug. In 2015, elucidation of the effectiveness of artemisinin as a potent anti-malarial drug was acknowledged with a Nobel prize. Owing to the tight market and low yield of artemisinin, an economical way to increase its production is to increase its content in Artemisia sp. through different biotechnological approaches including genetic transformation.
Methods
Artemisia annua and Artemisia dubia were transformed with rol ABC genes through Agrobacterium tumefacienes and Agrobacterium rhizogenes methods. The artemisinin content was analysed and compared between transformed and untransformed plants with the help of LC–MS/MS. Expression of key genes [Cytochrome P450 (CYP71AV1), aldehyde dehydrogenase 1 (ALDH1), amorpha-4, 11 diene synthase (ADS)] in the biosynthetic pathway of artemisinin and gene for trichome development and sesquiterpenoid biosynthetic (TFAR1) were measured using Quantitative real time PCR (qRT-PCR). Trichome density was analysed using confocal microscope.
Results
Artemisinin content was significantly increased in transformed material of both Artemisia species when compared to un-transformed plants. The artemisinin content within leaves of transformed lines was increased by a factor of nine, indicating that the plant is capable of synthesizing much higher amounts than has been achieved so far through traditional breeding. Expression of all artemisinin biosynthesis genes was significantly increased, although variation between the genes was observed. CYP71AV1 and ALDH1 expression levels were higher than that of ADS. Levels of the TFAR1 expression were also increased in all transgenic lines. Trichome density was also significantly increased in the leaves of transformed plants, but no trichomes were found in control roots or transformed roots. The detection of significantly raised levels of expression of the genes involved in artemisinin biosynthesis in transformed roots correlated with the production of significant amounts of artemisinin in these tissues. This suggests that synthesis is occurring in tissues other than the trichomes, which contradicts previous theories.
Conclusion
Transformation of Artemisia sp. with rol ABC genes can lead to the increased production of artemisinin, which will help to meet the increasing demand of artemisinin because of its diverse pharmacological and anti-malarial importance.
Similar content being viewed by others
Background
Artemisinin is a secondary metabolite that has been found to have strong anti-malarial activity with little or no side effects [1]. Artemisinin is a sesquiterpene lactone which has been reported to be produced within the glandular trichomes of Artemisia annua, a member of the Asteraceae family. This plant is endemic to China and has been used there for over 2000 years to treat fever [2, 3]. Artemisinin has also been found in aerial parts of Artemisia apiacea, Artemisia lancea and Artemisia cina [3–7] have also reported the presence of artemisinin in Artemisia dubia, Artemisia absinthium and Artemisia indica.
At present, artemisinin and its derivatives are used in combination therapies for the treatment of malaria [8], and for the treatment of a number of cancers and viral diseases [9, 10]. However, these therapies are not available to millions of the world’s poorest people because of the low yield of artemisinin in naturally growing Artemisia plants (0.1 to 0.5 % of dry weight) [11]. Mannan et al. [12] reported the presence of artemisinin in 17 naturally growing species of Artemisia and found artemisinin content in the range of 0.03 to 0.44 %. Enhanced production of artemisinin is, therefore, highly desirable. Genetic improvement of natural varieties has been attempted, but the maximum yield of artemisinin achieved so far is 2 % [13]. Even though there are some recent reports about cost effective chemical synthesis of artemisinin [14] or through microbial system [15], these methods are not yet practical for large-scale production of artemisinin. Due to complexity in production [8, 16], biosynthesis of artemisinin via the plant is still the preferred choice. If yields could be improved significantly, the market price could be stabilized and the drug made available at lower cost. One approach is to optimize production though modification of the metabolic pathways involved, this could be through selection for beneficial alleles or through genetic transformation.
Several groups have shown that transformation with different genes can increase artemisinin production through modification of its biosynthetic pathway. Overexpression of FPS in Artemisia annua increases the accumulation of artemisinin through conversion of IPP and DMADP into FDP (Fig. 1) [17]. Biswajit et al. [18] reported an increased amount of artemisinin in Artemisia annua transformed with Agrobacterium tumefaciens wild-type nopaline strains. Sa et al. [19] transferred the Ipt gene into Artemisia annua and observed increased amount of artemisinin in transformed plants. Over the past decade, increased production of secondary metabolites has been shown in plant cells transformed with the rol A, B and C genes. The rol A protein has emerged as a stimulator of growth and secondary metabolism [20, 21]. The rol B protein on the other hand, has been shown to have tyrosine phosphatase activity and, therefore, may play a role in the auxin signal transduction pathway [22]. Estruch et al. [23] demonstrated that rol C can be involved in the release of active cytokinins from their inactive glucosides due to its cytokinin glucosidase activity. rol C has also been shown in transformed plants and plant cell cultures to be capable of stimulating the production of tropane alkaloids [24], pyridine alkaloids [25], indole alkaloids [26], ginsenosides [27] and anthraquinones [28, 29].
Biosynthetic pathway of artemisinin. Figure showing pathway and key genes and enzymes involved in artemisinin biosynthesis [17]. ADS-Amorpha-4, 11-diene synthase, ALDH- Aldehyde dehydrogenase, CYP-CYP71AV1 (p450 enzymes), Dbr2 -Artemisinic aldehydeD11 (13) double bond reductase, DMAPP- Dimethyl allyl diphosphate, IPP- Isopentenyl diphosphate
The first step in artemisinin biosynthesis (Fig. 1) is the generation of amorpha-4, 11-diene by the cyclization of farnesyl diphosphate (FDP), catalyzed by amorpha-4, 11-diene synthase (ADS) [30]. Subsequent oxidation at the C12 position, mediated by the cytochrome P450 enzyme CYP71AV1, leads to artemisinic alcohol [31, 32]. While arteannuin B has been suggested as a late precursor in artemisinin biosynthesis [33, 34], evidence now favours a route from artemisinic alcohol via dihydroartemisinic acid [2, 35, 36].
Cloning and characterization of double bond reductase 2 (DBR2) supported this route, which reduces the D11 (13) double bond of artemisinic aldehyde, but not of arteannuin B [34], and the cloning of aldehyde dehydrogenase 1 (ALDH1), which catalyses the oxidation of artemisinic and dihydroartemisinic aldehyde [32]. According to Brown et al. [37–39], the conversion of dihydroartemisinic acid to artemisinin, and of artemisinic acid to arteannuin B occur via enzyme-independent reactions.
A number of publications have reported large differences in artemisinin content depending on variety, season, different plant parts, co cultivation conditions and plant developmental stage [30, 40–45]. Artemisinin has been detected in leaves; small green stems, buds, flowers and seeds [40, 45–47]. Reports on the distribution of artemisinin throughout the plant are however inconsistent. In some studies artemisinin has been reported to be higher at the top of the plant while others suggest it is equally distributed [48]. Artemisinin has also been shown to be produced by differentiated shoot cultures [46, 49, 50], but in shoots without roots, only trace levels are found, suggesting a regulatory effect [49–53]. Most workers [49, 50, 54, 55] have been unable to detect artemisinin in roots, although Nair et al. [56] and Tawfik et al. [54] reported trace amounts.
The current understanding is that artemisinin is produced in ten-celled glandular trichomes located on leaves, floral buds, and flowers [40, 57, 58] and sequestered in the epicuticular sac at the apex of the trichome [58]. For instance, artemisinin concentrations are higher in leaves that are formed later in development than in leaves formed early in the plant’s development; this difference has been attributed to a higher trichome density and a higher capacity per trichome in the upper leaves [59]. Artemisinin was thought not to be synthesized in roots [40] or pollen. ELISA analysis has shown that green roots can accumulate artemisinin (0.001 % dry weight), which was confirmed by GC–MS analysis [60]. Nguyen et al. [16] have also confirmed the presence of artemisinin in roots. It has also been shown that hairy roots produced by infection with Agrobacterium rhizogenes can produce artemisinin [60, 61]. Weathers et al. [62] reported levels of artemisinin (0.4 %) comparable to that found in the leaves within hairy root cultures of Artemisia annua transformed with Agrobacterium rhizogenes. This would suggest that the plant may be capable of producing artemisinin in the absence of trichomes.
Although the artemisinin concentration appears to be greatest in Artemisia annua, there are reports describing its presence in Artemisia absinthium, Artemisia dubia and Artemisia indica [5–7]. Chemical synthesis of artemisinin is difficult and, therefore, other Artemisia species or alternative sources, such as plant callus [63], shoot cultures [49] and hairy root cultures [64] offer attractive alternatives. These could be grown throughout the year, distancing production from time of harvest. Mannan et al. [7] found that Agrobacterium rhizogenes transformation increased the artemisinin content within hairy roots of Artemisia dubia to 0.6 % and to 0.23 % in Artemisia indica. Artemisia annua is restricted to certain global regions due to its growth requirements. Artemisia dubia is capable of growth in more varied conditions and could offer an alternative for production, ensuring security of production which would help ensure a constant price.
Numerous investigations have shown that the Agrobacterium rol genes can induce high levels of secondary metabolites in hairy root cultures of most transformed plant species. Data have been published about the effect of individual rol A, B and C genes on enhanced production of anti-malarial artemisinin [65–68].
The objective of the current study was the assessment of artemisinin production in transformed plants and hairy roots of Artemisia dubia and Artemisia annua transformed with rol ABC genes using Agrobacterium tumefaciens and Agrobacterium rhizogenes and their comparison with non-transformed plants. Trichome densities and the gene transcripts involved in the synthesis pathway were monitored to ascertain how the rol genes act to influence the production of anti-malarial artemisinin.
Methods
Seed sterilization and germination
Artemisia annua and Artemisia dubia seeds from a Northern Pakistan collection were disinfected with 70 % (v/v) ethyl alcohol for 30 s; surface sterilized with 0.1 % (w/v) Mercuric chloride (HgCl2) solution for 2 min, washed 3–4 min with sterile distilled water and transferred to half MS medium [69] containing 3 % (w/v) sucrose solidified with 0.8 % (w/v) agar. The seedlings were grown in growth room at 26 °C ± 2 with 16/8 h light dark cycle and light intensity maintained at 1000 lux.
Bacterial strains
Agrobacterium tumefaciens strain LBA4404 containing pRT99 was grown overnight in 50 ml of liquid malt yeast broth (MYB; 5 g/l yeast extract, 0.5 g/l casein hydrolysate, 8 g/l mannitol, 2 g/l ammonium sulfate, 5 g/l sodium chloride, pH 6.6). Agrobacterium rhizogenes strains LBA 9402 and 8196 containing Ri plasmid were grown overnight in 50 ml of liquid yeast mannitol broth (YMB; 0.5 g/l Di-potassium hydrogen phosphate, 2 g/l magnesium sulphate, 0.4 g/l yeast extract, 10 g/l mannitol, 0.1 g/l sodium chloride, pH 7.0). Prior to infection, the bacterial strains were grown overnight in liquid medium at 28 °C on a rotary shaker at 100 rpm in the dark.
Agrobacterium tumefaciens mediated transformation
Approximately 0.5–1 cm long stem and leaf explants were excised from the in vitro grown seedlings and were cultured on plain MS medium [69]. After 2–3 days of preculturing, these explants were dipped in Agrobacterium tumefaciens culture containing 50 mg/L kanamycin with different time durations; blotted on sterilize filter paper and cocultivated on MS medium containing 200 µM acetosyringone and 0.8 % agar.
After 3 days of cocultivation, the infected explants were washed with cefotaxime solution (500 µg/L), dried and transferred to selection MS medium containing 0.1 mg/L BAP (benzyl aminopurine), 20 mg/L kanamycin and 500 mg/L cefotaxime. These explants were maintained in growth room at 26 °C ± 2 with 16 h of photoperiod, illumination of 45 µE m−2s−1 and 60 % relative humidity. The explants were then transferred to fresh selection medium weekly during the 1 st month. Afterwards, subculturing was carried out every 2 weeks. After 8 weeks, the concentration of cefotaxime was reduced to 50 mg/L.
For rooting, the developed shoots cut off segments of Artemisia dubia were cultured on half MS medium containing 0.025 mg/L naphthalene acetic acid (NAA) solidified with 0.1 % gelrite and developed shoots cut off segments of Artemisia annua were cultured on MS medium containing 0.1 mg/L naphthalene acetic acid (NAA) solidified with 0.1 % gelrite.
Agrobacterium rhizogenes mediated transformation
Hairy roots induction
The four-week old plants of Artemisia dubia and Artemisia annua were infected with Agrobacterium rhizogenes strain LBA9402 and LBA8196. The stem portions were infected with 24 h old single colony of Agrobacterium rhizogenes with the help of scalpel. After production of hairy roots, 3–4 cm root tips were cut and placed on solid B5 medium for further proliferation.
Molecular analysis
For molecular analysis, genomic DNA from both Artemisia species was isolated by using CTAB method of [70] from transformed and untransformed plants, and plasmid DNA was also isolated by using alkaline lysis method from both Agrobacterium tumefaciens and Agrobacterium rhizogenes. PCR analysis was performed using a programmed DNA thermal cycler (Biometra, USA). The rol A gene forward 5′-AGAATGGAATTAGCCG GACTA-3′ and reverse primer 5′-GTATTAATCCCGTAGGTTTGTT-3′, the rol B forward 5′-GCTCTTGCAGTGCTAGATTT-3′ and reverse primer 5′-GAAGGTGC AAGCTACCT CTC-3′, the rol C gene forward 5′-GAAGACGACCTGTGTTCTC-3′ and reverse primer 5′-CGTTCAAACGTTAGCCGATT-3′ were used for PCR analysis. The PCR reaction was carried out in 25 µl final reaction volume containing 50 ng DNA template with the following thermal cycling conditions: 35 cycles of 5 min at 94 °C, 1 min at 53–55 °C and 1 min at 72 °C. Agarose gel (1.5 % w/v) electrophoresis was carried out to analyse 10 µl aliquot of PCR product.
Southern blot analysis of PCR-positive plants was performed by DIG High Prime DNA Labeling and Detection Starter Kit II (Roche Cat. No. 11585614910) according to the manufacturer’s instructions. For Southern blotting, 20 µg of genomic DNA was digested with EcoRI and electrophoresed on 0.8 % agarose gel. The DNA was denatured by alkaline solution and transferred to positively charged nylon membranes according to the standard procedure of PCR [96]. Products of rol A gene from plasmid was used as the probe. The probe was labeled using digoxigenin (DIG)-11-dUTP with DIG High Prime DNA labelling reagents (Roche, Mannheim, Germany). Hybridization was carried out at 42–45 °C followed by immunological detection on X-ray film using CSPD substrate according to the manufacturer’s instructions.
Analysis of artemisinin content
Chemicals
Reference standard of artemisinin (98 %), n-Hexane, ethyl acetate, LC–MS grade 0.1 % formic acid in water and acetonitrile was obtained from Sigma-Aldrich (Dorset, UK). Purified water (~18 MΩ/cm) was dispensed from a Milli-Q system (Millipore, UK).
Plant samples
Shoots, roots and hairy roots of transformed and untransformed plants of Artemisia annua (A1, A2, A3, AC, AR1, AR2, AR3, ARC, AH1, AH2) and Artemisia dubia (D1, D2, D3, DC, DR1, DR2, DR3, DRC, DH1, DH2) were used for the analysis of artemisinin with the help of LC–MS/MS.
Preparation of analytical standards
A standard stock solution of 1 mg mL−1 of artemisinin in acetonitrile was prepared. The analytical standard was prepared as dilution from the stock in the mobile phase in the concentration range between 0.15–10 µg mL−1. Beta-artemether was used as internal standard (IS) at 5 µg mL−1.
Sample extraction and preparation
Extraction of plant samples (shoots, roots, hairy roots) was based on published protocols [71, 72] with a slight modification. Briefly, 10 mL of n-hexane containing 5 % v/v ethyl acetate was used to extract 1 g of biomass in a cold (0 °C) sonication bath (PUL 125, Kerry Ultrasonics, UK) operated at 50 Hz for 30 min. The solvent from the extract was removed in vacuo and the residue re-suspended in 2 mL acetonitrile, which was then filtered through a 0.2 µm syringe filter (Fisher, UK) to remove waxes and other un-dissolved components. An aliquot of the filtrate placed in an HPLC vial was dissolved in the mobile phase and internal standard added for LC–MS/MS analysis.
Liquid chromatography method
The analysis was performed on an Aquity liquid chromatography unit coupled to a tandem quadruple detector (TQD) (Waters Corp., Milford, MA, USA). The liquid chromatography setup consisted of a binary pump, a cooling auto-sampler set at 10 °C with an injection loop of 10 µL. The column temperature was set at 30 °C and a Genesis® Lightn C18 column (100 × 2.1 mm; 4 µm) (Grace, IL, USA) was used for the separation of the metabolites. The method by [73] was employed which consisting of a binary mobile phase of A: 0.1 % formic acid in water and B: 0.1 % formic acid in acetonitrile. Chromatographic separation was achieved on a linear gradient run of 0–7.0 min, 25–98 % B; 7–9.5 min, 98 % B; 9.5–10 min, 98–25 % B; 10–15 min, 25 % B; and a flow rate of 0.4 mL min−1.
Multiple reaction monitoring (MRM) method
The mass spectrometer was operated in MRM mode with positive electrospray ionization (ESI+) using an earlier method [73]. Briefly, the cone and de-solvation gas flow rates were set at 45 L h−1 and 800 L h−1, respectively, while the capillary voltage, the source and de-solvation temperatures were similar for all analytes at 28 kV, 150 and 350 °C, respectively. Cone and collision voltages were set at 24 and 7 V, respectively. A MRM transition of 283 → 219 + 229 + 247 + 265 was used for identification and quantification of artemisinin. The dwell time was automatically set at 0.161 s. Data were acquired by MassLynx V4.1 software and processed for quantification with QuanLynx V4.1 (Waters Corp., Milford, MA, USA).
Analysis of metabolic pathway
Metabolic pathway of artemisinin production in Artemisia annua and Artemisia dubia and the effect of rol genes through which these genes enhance the production of artemisinin were analysed using different genes involved in artemisinin production, such as CYP71AV1 (Cytochrome P450), ADS (Amorpha-4, 11-diene, synthase), ALDH1 (Aldehyde dehydrogenase 1) and TFAR1 (Trichome-Specific fatty acyl-CoA reductase 1) involved in trichome development and sesquiterpenoid biosynthesis.
Quantitative real time PCR (qRT-PCR)
RNA extraction was performed using RNAqueous Small Scale Phenol-Free Total RNA Isolation Kit (Ambion) for small scale RNA isolation according to the manufacturer’s instructions. Frozen plant tissue powder (70 mg) was used for each RNA extraction. RNA (1 µg) was reverse transcribed using ThermoScript RT-PCR system (Invitrogen) primed with oligo (dT) 20primer. The RNA was removed from the first strand cDNA by RNase treatment using RNase H (Invitrogen) according to the manufacturer’s instructions.
The qRT-PCR was performed by using following specific primers (Table 1). Primers were designed for the five genes using PrimerSelect (Lasergene; DNAStar) based on the cDNA sequences specific for the Artemisia annua available at the National Centre for Biotechnology Information. Primer pairs were designed with almost similar melting temperatures, to amplify 200–300-bp fragments, and checked for amplification and specificity by gel electrophoresis of PCR products.
The real-time RT-PCR analysis was performed in an LightCycler® 480 Real-Time PCR System (Roche Applied Sciences, Foster City, CA, USA), using the cDNAs as templates and the random primers for the analysed gene, as listed in Table 1. The actin gene was used as a reference gene. SYBR Green (SYBR Premix Ex Taq; TaKaRa) was used in the polymerase chain reaction (PCR) to quantify the amount of dsDNA. PCR was performed using 3 µl of the cDNA in a total of 20 µl reaction volume. The PCR conditions were 2 min at 95 °C, 30 s at 95 °C, 30 s at 60–62 °C, 1 min at 72 °C for 45 cycles, followed by 5 min at 72 °C. These conditions were chosen because none of the samples analysed reached at exponential phase at the end of the amplification. Expression analysis of each gene was confirmed in at least three independent RT-reactions using forward and reverse primers.
Analysis of Trichome density
The number of glandular trichomes was determined at the adaxial side of three random pieces of fresh leaf material from each sample with an accurately determined leaf area of approximately 5 mm2 per leaf. Glandular trichomes were counted by using confocal microscope (Leica TCS SP5; Broadband Confocal, Leica Microsystems).
Each leaf was placed on a glass slide with three drops of Type F immersion liquid (Leica Microsystems CMs GmbH) with silicon vacuum grease at each corner below the cover slip to level the leaf. Clear nail polish sealed the cover slip to secure the sample and prevent evaporation. Using the confocal microscope three pictures of each leaf were taken and trichome density was analysed.
Statistical analysis
Experiments were independently replicated at least three times and significant differences in artemisinin content, expression of artemisinin synthesis pathway genes and trichome density were evaluated using Welch’s two-sample t test (Welch, 1947).
Results
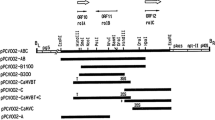
Three independent transgenic lines of Artemisia annua (A1, A2, and A3) and Artemisia dubia (D1, D2 and D3) were obtained by transformation with Agrobacterium tumefacienes and two independent transgenic lines of hairy roots were obtained by transformation with Agrobacterium rhizogenes (AH1 and AH2 of Artemisia annua and DH1 and DH2 of Artemisia dubia) carrying rol ABC genes (Fig. 2).
Comparison of transformed and untransformed plant transformed by Artemisia tumefaciens. A1–A3 represents the transformed plants of Artemisia annua; D1–D3 represents the transformed plants of Artemisia dubia. AC and DC are untransformed control plants of Artemisia annua and Artemisia dubia, respectively. AH represents the production of hairy roots from the plants of Artemisia annua; DH represents the production of hairy roots from the plants of Artemisia dubia
Molecular analysis
Molecular analysis of transformed plants/hairy roots was done with the help of PCR for rol A, B and C genes and the amplified products (308 bp, 779 bp, 540 bp respectively) were observed to confirm transformation. Same size amplified product was also obtained from the plasmid DNA of Agrobacterium strains. Rol A, B and C genes were detected in all transformed lines of shoots (A1, A2, A3, D1, D2, D3) roots (AR1, AR2, AR3, DR1, DR2, DR3) and hairy roots (AH1, AH2, DH1, DH2) as well as in plasmid DNA but not in control roots (Fig. 3a, b). Hybridization bands were detected in Southern blots with the rol A probe in the transgenic plants and hairy roots. In all the transformants, the inserted copy number was one except transgenic lines A1 and A3 of Artemisia annua and D2 of Artemisia dubia in which two copies of the inserted gene were observed (Fig. 4a, b). The results confirmed the integration of the Agrobacterium T-DNA in the genome of Artemisia annua and Artemisia dubia transgenic lines.
PCR analysis showing amplified products (a) A1–A3 represents the plants transformed by Agrobacterium tumefaciens and AH1–AH2 represents the plants transformed by Agrobacterium rhizogenes in Artemisia annua (b) D1–D3 represents the plants transformed by Agrobacterium tumefaciens and DH1–DH2 represents the plants transformed by Agrobacterium rhizogenes in Artemisia dubia. Lane P represents the plasmid DNA. Lane C refers to the non transformed control plants. Lane M corresponds to 1 kbp Ladder (Fermentas)
Southern blot analysis of (a) Artemisia annua (b) Artemisia dubia. C untransformed control plant, lanes 1–5 rol ABC transformed plants (Artemisia annua), lanes 1–5 rol ABC transformed plants (Artemisia dubia), A1–A3 represents the plants transformed by Agrobacterium tumefaciens and AH1-AH2 represents the plants transformed by Agrobacterium rhizogenes (Artemisia annua), D1-D3 represents the plants transformed by Agrobacterium tumefaciens and DH1–DH2 represents the plants transformed by Agrobacterium rhizogenes (Artemisia dubia)
Morphological characters
Considerable variation in the morphology of transgenic lines was observed as compared to controlled plants, like decrease in plant height with small and narrow leaves, terminal inflorescence and hard texture. The detailed morphological differences studied are listed in Table 2.
Analysis of artemisinin content
The average artemisinin content from three transgenic lines of Artemisia annua varied between tissues, the difference between the lines is shown (Fig. 5). In the shoots this was 33.12 mg/g dry weight (DW) compared to 3.94 mg/g DW in the controls shoots. Roots of transformed plants had an average artemisinin content of 2.72 mg/g DW while the amount found in transformed hairy roots was significantly greater (12.45 mg/g DW) compared to the control roots in which negligible amounts could be detected (0.02 mg/g DW). The average artemisinin in transgenic lines of Artemisia dubia was lower than Artemisia annua. Transgenic shoots of Artemisia dubia contained 2.32 mg/g DW compared to control shoots (0.11 mg/g DW), transformed plant roots (0.91 mg/g DW) and hairy roots (1.48 mg/g DW), whereas no artemisinin was found in control roots of Artemisia dubia.
Artemisinin content in shoots, roots and hairy roots of transformed and un-transformed plants of Artemisia dubia and Artemisia annua. (a) Artemisinin content in shoots roots and hairy roots of Artemisia annua. A1, A2 and A3 represents leaf samples from three different transgenic lines, AR1, AR2 and AR3 represents roots of transgenic lines A1, A2 and A3 respectively, AH1 and AH2 represents two transgenic lines of hairy roots, AC and ARC represent control shoots and roots. (b) Artemisinin content in shoots, roots and hairy roots of Artemisia dubia. D1, D2 and D3 represent leaves from three transgenic lines. DR1, DR2 and DR3 represents transformed roots of transgenic lines D1, D2 and D3 respectively, DH1 and DH2 represents two transgenic lines of hairy roots, DC and DRC represent control shoots and roots. Both technical and biological replicates were performed. The average of three plants is shown together with error bars showing SD. Data was analyded for significant difference compared to the controls and all showed P < 0.001 with the control by Welch’s two samples t test
Analysis of metabolic pathway and trichome development
The genes ADS, CYP71AV1 and ALDH1 are involved in the metabolic synthesis pathway of artemisinin and are involved in the conversion of FDP to dihydroartemisinic acid, which is a late precursor of artemisinin (Fig. 1). These key genes were used to determine how transformation with the Agrobacterium rol ABC genes affected the pathway in the different tissues of Artemisia annua and Artemisia dubia. Trichome-Specific fatty acyl-CoA reductase 1 (TAFR1) has been suggested to be involved in trichome development and sesquiterpenoid biosynthesis, both of which are important for artemisinin production. The expression of this gene was monitored to determine if trichome density might also be affected. Figure 6 shows expression levels of these genes in different tissues of both transformed and untransformed Artemisia annua and Artemisia dubia.
Expression of key genes CYP71AV1 (Cytochrome P450), ADS (Amorpha-4, 11-diene, synthase), ALDH1 (Aldehyde dehydrogenase 1) involved in artemisinin synthesis pathway of Artemisia annua and Artemisia dubia for artemisinin biosynthesis, and TFAR1 (Trichome-Specific fatty acyl-CoA reductase 1) involved in trichome development and sesquiterpenoid biosynthesis. a. Expression of genes in leaves of Artemisia annua. A1, A2 and A3 represents three different transgenic lines of leaves, AC represents control shoots. b. Expression of genes in leaves of Artemisia dubia. D1, D2 and D3 represents three different transgenic lines of leaves, DC represents control shoots. c. Expression of genes in hairy roots, roots of transformed shoots of Artemisia annua. AH1 and AH2 represent two transgenic lines of hairy roots; AR1, AR2 and AR3 represent transformed roots of transgenic lines A1, A2 and A3 respectively. ARC represents control roots. d. Expression of genes in hairy roots, roots of transformed shoots of Artemisia dubia. DH1 and DH2 represent two transgenic lines of hairy roots; DR1, DR2 and DR3 represent transformed roots of transgenic lines D1, D2 and D3 respectively. DRC represents control roots. The actin gene was used as a house-keeping control. Both technical and biological replicates were performed. The average of three plants is shown together with error bars showing SD. Data analysed for significant difference at P < 0.001 with the control by Welch’s two samples t-test
Relative expression of genes involved in metabolic pathway of artemisinin biosynthesis
Pronounced changes in the expression of the artemisinin biosynthetic pathway genes were observed in all transgenic lines of Artemisia annua and Artemisia dubia transformed with rol ABC genes (Fig. 6a–d). The qPCR data presented here clearly demonstrate that the expression levels of ADS, CYP71AV1 and ALDH1 were significantly increased (P < 0.0001) in transformed plants of both Artemisia sp. compared to untransformed plants. Expression of artemisinin biosynthetic pathway genes was observed in hairy roots and roots of transformed plants compared to control roots of both Artemisia annua and Artemisia dubia in which low or negligible levels of expression are found. Expression levels of all the artemisinin biosynthetic genes were also significantly higher in transformed shoots compared to control plants. Interestingly, transformed Artemisia dubia showed levels, which were greater than, control Artemisia annua plants despite similar artemisinin content. Not all the genes examined exhibited the same increase in expression. Cyp71AV1 appeared to show the greatest changes in relative expression in both shoots and hairy roots of both Artemisia annua and Artemisia dubia compared to the other genes. Relative expression of CYP71AV1gene was higher than that of the other genes. Its expression was significantly increased in transformed plants of both Artemisia species compared to untransformed plants, i.e. ~5 and ~120 times more in transformed leaves of Artemisia annua and Artemisia dubia, respectively. Roots of transformed plants in both Artemisia species showed significantly higher expression of this gene compared to control roots. The greatest difference in expression of this gene was found in hairy roots from both species even compared to transformed roots and control roots.
Relative expression of the ADS gene was generally lower than that of the other genes however the difference in its expression compared to the control was the highest. i.e. ~11 and ~270 greater in transformed leaves of Artemisia annua and Artemisia Dubia, respectively. Roots from these plants showed even larger differences in the expression of this gene compared to the controls. In Artemisia annua, the levels of expression of ADS was comparable between transformed roots and hairy roots, but in Artemisia dubia there was a significant difference in the expression of this gene.
The ALDH1 gene showed high expression in transformed plants of both Artemisia species when compared to untransformed plants, i.e. ~7 and ~5 times more in transformed leaves of Artemisia annua and Artemisia dubia, respectively. Roots of these transformed plants also showed greater differences in expression of this gene in Artemisia annua compared to control, in contrast in Artemisia dubia the level of expression of this gene was comparatively low. However expression of this gene showed significant differences in hairy roots of both Artemisia species compared to control roots.
Trichome development
A significant increase in expression of TFAR1 was detected in transformed plants of both Artemisia species compared to untransformed plants, i.e. ~10 and ~300 times more in transformed leaves of Artemisia annua and Artemisia dubia, respectively. Unexpectedly this gene was expressed in abundance in transformed roots and hairy roots of both Artemisia species. The levels of expression were greater than that seen in control shoots. Although this gene has been suggested to be responsible for trichome density it showed significant differences in expression even in transformed roots and hairy roots compared to control roots.
Analysis of trichome density
Trichome density was assessed using ESEM at low resolution and is shown in Fig. 7. Transformed leaves of Artemisia annua plants produced more trichomes (~222 trichomes/5 mm2) as compared to the control leaves (~120 trichomes/5 mm2). Transformed leaves of Artemisia dubia also produced more trichomes (~173 trichomes/5mm2) compared to control leaves (~173 trichomes/5mm2) In contrast, the hairy roots, transformed and untransformed roots from both Artemisia species showed no trichome production (Figs. 7, 8).
Comparison of trichome density in different tissues of transformed and untransformed plants of Artemisia annua and Artemisia dubia. Trichome density was analysed by using confocal microscope. A1, A2 and A3 represents three transgenic lines of leaves in Artemisia annua, AH represents hairy roots; AC and ARC represent control shoots and roots of Artemisia annua, respectively. D1, D2 and D3 represent three transgenic lines of leaves in Artemisia dubia, DH represents hairy roots, DC and DRC represent control shoots and roots of Artemisia dubia, respectively. Both technical and biological replicates were performed
Comparative analysis of trichome density in transformed and untransformed plants of Artemisia annua and Artemisia dubia. a Trichome density in shoots roots and hairy roots of Artemisia annua. A1, A2 and A3 represents three different transgenic lines of leaves, AR1, AR2 and AR3 represents transformed roots of transgenic lines A1, A2, A3 respectively, AH1 and AH2 represents two transgenic lines of hairy roots, AC and ARC represent control shoots and roots of respectively. b. Trichome density in shoots roots and hairy roots of Artemisia dubia. D1, D2 and D3 represents three transgenic lines of leaves, DR1, DR2 and DR3 represents transformed roots of transgenic lines D1, D2, D3, respectively, DH1 and DH2 represents two transgenic lines of hairy roots, DC and DRC represent control shoots and roots of respectively. Both technical and biological replicates were performed. The average of three plants is shown together with error bars showing SD. Data analysed for significant difference at P < 0.001 with the control by Welch’s two samples t test
Transformation also increased hairiness in transformed plants along with increase in trichome density compared to non-transformed plants. Transformed plants produced denser network of hair on its surface compared to non-transformed plants (Fig. 7).
Discussion
Artemisinin analysis was carried out on dried plant material (shoots, roots and hairy roots) of transformed and non-transformed plants of Artemisia annua and Artemisia dubia. The tissue assayed was similar to that from Artemisia species in previous studies [5, 41, 74–77]. Control plants in the current study contained similar amounts of artemisinin when compared to these studies on the vegetative stage. In the present study, 0.39 % DW artemisinin in control plants of Artemisia annua and 0.01 % DW artemisinin in Artemisia dubia, while Wallaart et al. [11] found 0.1–0.5 % DW artemisinin in control plants of Artemisia annua, Weathers et al. [78] found 0.2–0.5 % artemisinin in vegetative leaves and Mannan et al. [72] found 0.44 % DW artemisinin in Artemisia annua leaves and 0.05 % DW in leaves of Artemisia dubia.
Transformed shoots, roots of transformed shoots and hairy roots of Artemisia annua and Artemisia dubia all contained significantly more artemisinin when compared to the controls. The highest concentration of artemisinin were observed in transgenic leaves of both Artemisia species (i.e. ~9 and ~25 fold increase in leaves of Artemisia annua and Artemisia dubia, respectively, compared to controls). Other researchers have previously reported that transformation of different Artemisia species results in an increase in production of artemisinin within the plant. Vergauwe et al. [79] reported that Agrobacterium tumefacienes transformation with EHA101 (pTJK136) strain and C58C1RifR (pGV2260) (pGSJ780BAT) strain of Artemisia annua resulted in an increase in artemisinin content (0.42 %) and its metabolites. He et al. [63] also reported 0.36 % increase in artemisinin amount in transformed leaves of Artemisia annua as compared to non-transformed plants. Similarly, Adbin et al. [47] reported the presence of artemisinin, ranging from 0.01 to 0.80 %, in leaves and flowers of transformed Artemisia annua and by overexpression of genes coding for enzymes associated with the rate limiting steps of artemisinin biosynthesis pathway. Han et al. [80] reported that artemisinin production increased by up regulation of FDS through genetic transformation of Artemisia annua with different genes. Overexpression of Gossypium arboretum FPS in hairy roots and transgenic plants resulted in three- to four-fold, and two- to three-folds increased artemisinin content, respectively [81, 82]. The results achieved in this study are significantly higher, with increase of 12-fold or greater being detected depending on the tissue examined.
A meaningful increase in artemisinin content was observed in hairy roots and roots of transformed shoots of Artemisia annua and Artemisia dubia as compared to control, similarly roots of transformed plants in both Artemisia species also showed significant increase in artemisinin content compared to control roots. An earlier report shows that Agrobacterium rhizogenes mediated transformation increased the level of secondary metabolites in plants [83]. Artemisinin production (0.42 %) has been reported in hairy roots culture of Artemisia annua [62]. Mannan et al. [7] reported the production of artemisinin by hairy roots in Artemisia dubia (0.603 %) and Artemisia indica (0.0026 %) with Agrobacterium rhizogenes strain LBA9402. A significant increase in artemisinin content in hairy roots culture of Artemisia Annua was also reported [84]. The concentration of artemisinin in roots of untransformed Artemisia annua and Artemisia dubia is almost negligible (Fig. 5a, b). These findings agree with others [50, 85] who found very low artemisinin concentrations (0.003 %) in root of Artemisia annua. Other studies did not detect artemisinin within roots of Artemisia annua perhaps due to the sensitivity of the analysis carried out [49, 52, 55, 63, 86]. The current study also found low (Artemisia annua) or zero (Artemisia dubia) artemisinin concentration in untransformed roots. Transformed plants, hairy roots and roots of transformed shoots of Artemisia dubia and Artemisia annua were all found to contain significantly higher amounts of artemisinin.
The results obtained clearly show that the roots of transformed plants and hairy roots both contain around 1.5 % artemisinin. This is only slightly less than that found in leaves of Artemisia annua and significantly higher than that found in leaf material from Artemisia dubia. It has been reported that exogenous artemisinin is capable of inhibiting root growth at concentrations greater than 10 μg/ml−1 [87]. The study showed that hairy roots were capable of producing more than 15 μg g−1 DW. This would suggest that there are certain cells within the roots which are acting as storage compartments for the artemisinin produced. Mannan et al. [12] had shown that roots of Artemisia plants are capable of producing artemisinin. Interestingly the roots do not contain trichomes; hair cells are however present and it is possible that these cells are also involved in the production and storage of artemisinin. The number of hair cells also appeared to be present in greater abundance on the leaves of transformed plants (Fig. 7).
Presented study found that expression of all the genes involved in artemisinin synthesis pathway significantly increased in all transgenic lines of Artemisia dubia and Artemisia annua transformed with rol ABC genes as compared to non-transformed plants (Fig. 6a, b, c, d), which is in clear agreement with the data obtained for artemisinin content.
Expression of ADS and CYP71AV1, of which the corresponding proteins catalyze the formation of amorpha-4, 11-diene and its ultimate conversion to dihydroartemisinic acid, was increased significantly in transformed shoots of Artemisia annua and Artemisia dubia, respectively. Similarly significant differences in expression of these genes were observed in hairy roots and roots of transformed plants of both Artemisia annua and Artemisia dubia compared to untransformed roots. The expression of CYP71AVI corresponds to that found by [88] however the result for the ADS gene differs as they were unable to show ADS gene expression in hairy roots. The ALDH1 gene which catalyses the oxidation of artemisinic and dihydroartemisinic aldehydes was also highly expressed in transformed plants of both Artemisia species as compared to untransformed plants, i.e. ~7 and ~5 times more in transformed leaves of Artemisia annua and Artemisia dubia, respectively, ~5times in roots of transformed shoots of Artemisia annua and in Artemisia dubia this increased from negligible amounts to significant levels, with similar increases in hairy roots compared to untransformed roots (Fig. 6a–d). Although all these genes showed increased expression in transformed plants and hairy roots, however induction of CYP71AV1 and ALDH1 was much more compared to the ADS gene.
TFAR1, known to stimulate trichome development and to catalyze sesquiterpenoid biosynthesis [17] showed significantly increased expression in all transgenic lines of Artemisia annua and Artemisia dubia (Fig. 6). Density of trichomes were also measured in transformed and non-transformed plants and found that transformed leaves of Artemisia annua and Artemisia dubia have a higher density of trichomes compared to leaves of non-transformed plants. Despite close examination no trichomes were found on roots and hairy roots cultures of either Artemisia species but they were shown to contain significant amount of artemisinin as shown in Fig. 5a, b. Olofsson et al. [88] found that leaves and flower buds of Artemisia annua produce more trichomes than other parts of the plant and also found that roots do not have trichomes. Although the pathway for the synthesis of artemisinin is well known [89], it has been widely assumed that the trichomes are the only source of artemisinin production in the plant [40, 50, 57, 58] despite several papers describing its presence within roots [12, 51, 56, 60], reports that the artemisinin content of the plant is associated with a rise in trichome density, but also continued to rise after a collapse in the trichome populations. The results of present study found that the trichome density in Artemisia annua is increased by 10 % following transformation however the artemisinin content increased by approximately 10 times. In Artemisia dubia, trichome density was increased by 9 % following transformation and artemisinin content was increased by approximately 30 times (Figs. 5a, b, 7, 8). These results suggest a possible dissociation between trichome density and artemisinin content. It is unclear if the genes involved in artemisinin production are expressed elsewhere in the leaves [90]. It has been previously suggested that the plant was unable to support concentrations higher than this and that the trichomes capacity for storing artemisinin is limited [91]. The current study shows that the plants are capable of producing and storing much higher amounts of artemisinin than previously thought both in leaves and in roots. This study shows significant amount of artemisinin production in transformed roots of Artemisia annua and Artemisia dubia, and a small amount of artemisinin in untransformed roots of Artemisia annua. The detection of significantly raised levels of expression of the genes involved in artemisinin in transformed roots associated with the detection of significant amounts of artemisinin in these tissues suggest that synthesis is occurring elsewhere in the plant as suggested by Duke et al. [90]. It should now be possible to determine how roots produce artemisinin hence elucidating how production could be improved.
Baldi and Dixit [92] and Wang et al. [93] have reported that artemisinin production in plants and culture cells is increased after treatment with methyl Jasmonate; another report examined the effect of cytokinin, gibberelin and jasmonate on artemisinin production [17]. Similarly, Arsenault et al. [94] described the effect of sugars on the production of artemisinin in Artemisia annua. Current study deal with the transformation of Artemisia annua and Artemisia dubia plants with rol ABC genes of Agrobacterium tumefaciens and Agrobacterium rhizogenes, which are responsible for enhancing production of secondary metabolites in plants. The exact mechanism for the action of the rol genes is still far from clear [26], it has been suggested that they act through the stimulation of the plants defense response which includes induction of many of the hormonal pathways described above. Many secondary metabolites are thought to be have evolved to protect plants as they expanded to fill different evolutionary niches. It has been suggested that artemisinin is an example of such a metabolite as it acts to prevent predation of the plant by herbivores and it is stimulated by pruning prior to harvest [95]. This may explain why the transformation with Agrobacterium rol genes has such a significant effect on the amount of artemisinin produced by the transformed plants. Stimulation of the synthesis of artemisinin within these plants has also allowed us to alter the perception of how and where artemisinin is produced within the plants. These findings will help to develop better strategies to increase the production of this valuable therapeutic drug.
Conclusion
Transformation of Artemisia annua and Artemisia dubia with rol ABC genes results in the enhancement of secondary metabolites particularly the artemisinin and its derivatives. Moreover, the level of transcripts of the rol ABC genes gene found in transgenics also correlate with their artemisinin accumulation pattern. This report covers the impact these genes have had on the capability of the plants to synthesize the anti-malarial drug. Analysis of genes involved in the synthetic pathways and comparisons of trichome density between transformed and non-transformed plants have been carried out. Significantly, an increase in trichome numbers and increased expression of biosynthetic pathway genes correlated with increased artemisinin content. This data allow us to present a novel interpretation of artemisinin production and how stimulation of the pathways by the rol ABC genes can enhance artemisinin production stimulating both the biosynthetic pathways and trichome development in transformed plants of Artemisia dubia and Artemisia annua. The insight this study has made into how and where artemisinin is produced within the plants will provide vital information that is required to increase production of artemisinin to meet the increasing demand of drug manufacturers due to its multi pharmacological importance.
Abbreviations
- CYP71AV1 :
-
cytochrome P450
- ADS :
-
amorpha-4 11-64 diene synthase
- ALDH1 :
-
aldehyde dehydrogenase
- TFAR1 :
-
trichome-specific fatty acyl-CoA reductase 1
References
Klayman DL. Qinghaosu (artemisinin): an anti-malarial drug from China. Science. 1985;228:1049–55.
Covello PS. Making artemisinin. Phytochemistry. 2008;69:2881–5.
Hsu E. The history of Qinghao in the Chinese materiamedica. Trans R Soc Trop Med Hyg. 2006;100:505–8.
Aryanti BM, Ermayanti TM, Mariska I. Production of antileukemic agent in untransformed and transformed roots cultures of Artemisia cina. Annales Bogorienses. 2001;8:11–6.
Arab HA, Rahbari S, Rassouli A, Moslemi MH, Khosravirad FDA. Determination of artemisinin in Artemisia sieberi and anticoccidial effects of the plant extract in broiler chickens. Trop Anim Health Prod. 2006;38:497–503.
Zia M, Mannan A, Chaudhary MF. Effect of growth regulators and amino acids on artemisinin production in the callus of Artemisia absinthium. Pak J Bot. 2007;39:799–805.
Mannan A, Shaheen N, Arshad W, Qureshi RA, Zia M, Mirza B. Hairy roots induction and artemisinin analysis in Artemisia dubia and Artemisia indica. Afr J Biotechechnol. 2008;7:3288–92.
Haynes RK. From artemisinin to new artemisinin anti-malarials: biosynthesis, extraction old and new derivatives, stereochemistry and medicinal chemistry requirements. Curr Top Med Chem. 2006;6:509–37.
Efferth T. Antiplasmodial and antitumor activity of artemisinin from bench to bedside. Planta Med. 2007;73:299–309.
Efferth T, Romero MR, Wolf DG, Stamminger T, Marin JJG, Marschall M. The antiviral activities of artemisinin and artesunate. Clin Infect Dis. 2008;47:804–11.
Wallaart TE, Pras N, Quax WJ. Seasonal variations of artemisinin and its biosynthetic precursors in tetraploid Artemisia annua plants compared with the diploid wild-type. Planta Med. 1999;65:723–8.
Mannan A, Ahmed I, Arshad W, Asim MF, Qureshi RA, Hussain I, et al. Survey of artemisinin production by diverse Artemisia species in northern Pakistan. Malar J. 2010;9:310.
Graham IA, Besser K, Blumer S, Branigan CA, Czechowski T, Elias L, et al. The genetic map of Artemisia annua L. Identifies Loci affecting yield of the anti-malarial drug artemisinin. Science. 2010;327:328–31.
Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, et al. High-level semi-synthetic production of the potent anti-malarial artemisinin. Nature. 2013;496:528–32.
Zhu C, Cook SP. A concise synthesis of (+)-artemisinin. J Am Chem Soc. 2012;134:13577–9.
Nguyen KT, Towler MJ, Weathers PJ. The effect of roots and media constituents on trichomes and artemisinin production in Artemisia annua L. Plant Cell Rep. 2013;32:207–18.
Maes L, Van Nieuwerburgh FC, Zhang Y, Reed DW, Pollier J, Vande Casteele SR, et al. Dissection of the phytohormonal regulation of trichome formation and biosynthesis of the anti-malarial compound artemisinin in Artemisia annua plants. New Phytol. 2011;189:176–89.
Biswajit G, Swapna M, Sumita J. Genetic transformation of Artemisia annua by Agrobacterium tumefaciens and artemisinin synthesis in transformed cultures. Plant Sci. 1997;122:193–9.
Sa G, Mi M, He-Chun Y, Ben-Ye L, Guo-feng LKC. Effects of ipt gene expression on the physiological and chemical characteristics of Artemisia annua L. Plant Sci. 2001;160:691–8.
Altvorst AC, Bino RJ, Van Dijk AJ, Lamers AMJ, Van Den Mark F, Dons JJM. Effects of the introduction of Agrobacteirum rhizogenes rol genes on Artemisia plant and flower development. Plant Sci. 1992;83:77–85.
Schmulling T, Fladung M, Grossmann K, Schell J. Hormonal content and sensitivity of transgenic tobacco and potato plants expressing single rol genes of A. rhizogenes T-DNA. Plant J. 2001;3:371–82.
Filippini F, Rasi V, Marin O, Trovato M, Downey PM, Lo Shiavo F, et al. A plant oncogene as a phosphatase. Nature. 1994;379:499–500.
Estruch JJ, Schell J, Spena A. The protein encoded by the rol B plant oncogene hydrolysis indole glucosides. EMBO J. 1999;10:3125–8.
Bonhomme V, Laurain-Mattar D, Fliniaux MA. Effects of the rolC gene on hairy root induction development and tropane alkaloid production by Atropa belladonna. J Nat Prod. 2000;63:1249–52.
Palazón J, Cusidó RM, Roig C, Piñol MT. Expression of the rol C gene and nicotine production in transgenic roots and their regenerated plants. Plant Cell Rep. 1998;17:384–90.
Bulgakov VP. Functions of rol genes in plant secondary metabolism. Biotechnol Adv. 2008;26:318–24.
Bulgakov VP, Tchernoded GK, Mischenko NP, Khodakovskaya MV, Glazunov VP, Zvereva EV. Effects of salicylic acid, methyl jasmonate, etephone and cantharidin on anthraquinone production by Rubia cordifolia callus cultures transformed with rolBandrolC genes. J Biotechnol. 2002;97:213–21.
Bulgakov VP, Tchernoded GK, Mischenko NP, Shkryl YN, Glazunov VP, Fedoreyev SA. Effects of Ca2+ channel blockers and protein kinase/phosphatase inhibitors on growth and anthraquinone production in Rubia cordifolia cultures transformed by the rol B and rolC genes. Planta. 2003;217:349–55.
Shkryl YN, Veremeichik GN, Bulgakov VP, Tchernoded GK, Mischenko NP, Fedoreyev SA. Individual and combined effects of the rol A, B and C genes on anthraquinone production in Rubia cordifolia transformed calli. Biotechnol Bioeng. 2008;100:118–25.
Wallaart TE, Pras N, Beekman AC, Quax WJ. Seasonal variation of artemisinin and its biosynthetic precursors in plants of Artemisia annua of different geographical origin: proof for the existence of chemotypes. Planta Med. 2000;66:57–62.
Ro D-K, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, et al. Production of the anti-malarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–3.
Teoh KH, Polichuk DR, Reed DW, Nowak G, Covello PS. Artemisia annua L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the anti-malarial sesquiterpene lactone artemisinin. FEBS Lett. 2006;580:1411–6.
Sangwan RS, Agarwal K, Luthra R, Thakur RS, Singh-Sangwan N. Biotransformation of arteannuic acid into arteannuin-B and artemisinin in Artemisia annua. Phytochemistry. 1993;34:1301–2.
Zhang Y, Teoh KH, Reed DW, Maes L, Goossens A, Olson DJH, et al. The molecular cloning g of artemisinic aldehyde D11 (13) reductase and its role in glandular trichome-dependent biosynthesis of artemisinin in Artemisia annua. J Biol Chem. 2008;283:21501–8.
Bertea CM, Freije JR, van der Woude H, Verstappen FWA, Perk L, Marquez V, et al. Identification of intermediates and enzymes involved in the early steps of artemisinin biosynthesis in Artemisia annua. Planta Med. 2005;71:40–7.
Covello PS, Teoh KH, Polichuk DR, Reed DW, Nowak G. Functional genomics and the biosynthesis of artemisinin. Phytochemistry. 2007;68:1864–71.
Sy L-K, Brown GD. The mechanism of the spontaneous autoxidation of dihydroartemisinic acid. Tetrahedron. 2002;58:897–908.
Brown GD, Sy LK. In vivo transformations of dihydroartemisinic acid in Artemisia annua plants. Tetrahedron. 2004;60:1139–59.
Brown GD, Sy LK. In vivo transformations of artemisinic acid in Artemisia annua plants. Tetrahedron. 2007;63:9548–66.
Ferreira JFS, Simon JE, Janick J. Developmental studies of Artemisia annua: flowering and artemisinin production under greenhouse and field conditions. Planta Med. 1995;61:167–70.
Delabays N, Simonnet X, Gaudin M. The genetics of artemisinin content in Artemisia annua L. and the breeding of high yielding cultivars. Curr Med Chem. 2001;8:1795–801.
Lommen WJM, Elzinga S, Verstappen FWA, Bouwmeester HJ. Artemisinin and sesquiterpene precursors in dead and green leaves of Artemisia annua L. crops. Planta Med. 2007;73:1133–9.
Davies MJ, Atkinson CJ, Burns C, Woolley JG, Hipps NA, Arroo RRJ, et al. Enhancement of artemisinin concentration and yield in response to optimization of nitrogen and potassium supply to Artemisia annua. Ann Bot. 2009;104:315–23.
Yang RY, Zeng XM, Lu YY, Lu WJ, Feng LL, Yang XQ, et al. Senescent leaves of Artemisia annua are one of the most active organs for overexpression of artemisinin biosynthesis responsible genes upon burst of singlet oxygen. Planta Med. 2009;76:734–42.
Towler MJ, Weathers PJ. Variations in key artemisinic and other metabolites throughout plant development in Artemisia annua L. for potential therapeutic use. Ind Crops Prod. 2015;67:185–91.
Martinez BC, Staba J. The production of artemisinin in Artemisia annua L. tissue cultures. Adv Cell Cult. 1988;6:69–87.
Abdin MZ, Israr M, Rehman RU, Jain SK. Artemisinin, a novel anti-malarial drug: biochemical and molecular approaches for enhanced production. Planta Med. 2003;69:289–99.
Laughlin JC. The influence of distribution of anti-malarial constituents in Artemisia annua L. on time and method of harvest. Acta Hort. 1995;390:67–73.
Fulzele DP, Sipahimalani AT, Heble MR. Tissue cultures of Artemisia annua: organogenesis and artemisinin production. Phytother Res. 1991;5:149–53.
Ferreira JFS, Janick J. Roots as an enhancing factor for the production of artemisinin in shoot cultures of Artemisia annua. Plant Cell Tissue Organ Cult. 1996;44:211–7.
Jha J, Jha TB, Mahato SB. Tissue culture of Artemisia annua L.: a potential source of an anti-malarial drug. Curr Sci. 1988;57:344–6.
Woerdenbag HJ, Pras N, Bos R, Visser JF, Hendriks H, Malingre TM. Analysis of artemisinin and related sesquiterpenoids from Artemisia annua L. by combined gas chromatography/mass spectrometry. Phytochem Anal. 1991;2:215–9.
Paniego NB, Giuliette AM. Artemisia annua L.: dedifferentiated and differentiated cultures. Plant Cell Tiss Organ Cult. 1994;36:163–8.
Tawfiq NK, Anderson LA, Roberts MF, Phillipson JD, Bray DH, Warhurst DC. Antiplasmodial activity of Artemisia annua plant cell cultures. Plant Cell Rep. 1989;8:425–8.
Kim NC, Kim SU. Biosynthesis of artemisinin from 11,12-dihydroarteannuic acid. J Korean Agric Chem Soc. 1992;35:106–9.
Nair MSR, Acton N, Klayman DL, Kendrick K, Basile DV, Mante S. Production of artemisinin in tissue cultures of Artemisia annua. J Nat Prod. 1986;49:504–7.
Tellez MR, Canel C, Rimando AM, Duke SO. Differential accumulation of isoprenoids in glanded and glandless Artemisia annua L. Phytochemistry. 1999;52:1035–40.
Olsson ME, Olofsson LM, Lindahl AL, Lundgren A, Brodelius M, Brodelius PE. Localization of enzymes of artemisinin biosynthesis to the apical cells of glandular trichomes of Artemisia annua L. Phytochemistry. 2009;70:1123–8.
Lommen WJM, Schenk E, Bouwmeester HJ, Verstappen FWA. Trichome dynamics and artemisinin accumulation during development and senescence of Artemisia annua leaves. Planta Med. 2006;72:336–45.
Jaziri M, Shimonura K, Yoshimatsu K, Fauconnier ML, Marlier M, Homes J. Establishment of normal and transformed root cultures of Artemisia annua L. for artemisinin. J Plant Physiol. 1995;145:175–7.
Liu CZ, Wang YC, Zhao B, Guo C, Ouyang F, Ye HC, Li GF. Development of a nutrient mist bioreactor for growth of hairy roots in vitro. Cell Dev Biol Plant. 1999;35:271–4.
Weathers PJ, Cheetham RD, Follansbee E, Theoharides K. Artemisinin production by transformed roots of Artemisia annua. Biotechnol Lett. 1994;16:1281–6.
He XC, Zeng MY, Li GF, Liang Z. Callus induction and regeneration of plantlets from Artemisia annua and changes of qinghaosu contents. Acta Bot Sin. 1983;25:87–90.
Qin MB, Li GZ, Ye HC, Li GF. Induction of hairy root from Artemisia annua with Agrobacterium Rhizogenes and its culture in vitro. Acta Bot Sin. 1994;36:165–70.
Giri A, Narasu LM. Transgenic hairy roots: recent trends and applications. Biotechnol Adv. 2000;18:1–22.
Sevon N, Oksman-Caldentey KM. Agrobacterium rhizogenes-mediated transformation: root cultures as a source of alkaloids. Planta Med. 2002;68:859–68.
Dilshad E, Cusido RM, Palazon J, Estarada KR, Bonfil M, Mirza B. Enhanced artemisinin yield by expression of rol genes in Artemisia annua. Malar J. 2015;14:424.
Dilshad E, Cusido RM, Palazon J, Estarada KR, Bonfil M, Mirza B. Genetic transformation of Artemisia carvifolia Buch with rol genes enhances artemisinin accumulation. PLoS One. 2015;10:e0140266.
Murashige T, Skoog E. A revised medium for rapid growth and bioassay with tobacco tissue culture. Physiol Plant. 1962;15:472–93.
Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–5.
Lapkin AA, Walker A, Sullivan N, Khambay B, Mlambo B, Chemat S. Development of HPLC analytical protocols for quantification of artemisinin in biomass and extracts. J Pharm Biomed Anal. 2009;49:908–15.
Mannan A, Liu C, Arsenault PR, Towler MJ, Vail DR, Lorence A, Weathers PJ. DMSO triggers the generation of ros leading to an increase in artemisinin and dihydroartemisinic acid in Artemisia annua shoot cultures. Plant Cell Rep. 2010;29:143–52.
Suberu J, Gromski PS, Nordon A, Lapkin A. Multivariate data analysis and metabolic profiling of artemisinin and related compounds in high yielding varieties of Artemisia annua field-grown in Madagascar. J Pharm Biomed Anal. 2016;117:522–31.
Guo PQ, Yi-Wen Y, Qi-Long R. Determination of artemisinin in Artemisia annua L. by reversed phase HPLC. J Liquid Chromat Related Tech. 2005;28:705–12.
Wang M, Park C, Wu Q, Simon JE. Analysis of artemisinin in Artemisia annua L. by LC-MS with selected ion monitoring. J Agric Food Chem. 2005;53:7010–3.
Peng CA, Ferreira JF, Wood AJ. Direct analysis of artemisinin from Artemisia annua L. using high-performance liquid chromatography with evaporative light scattering detector and gas chromatography with flame ionization detector. J Chromat A. 2006;11:254–8.
Towler MJ, Weathers PJ. Evidence of artemisinin production from IPP stemming from both the mevalonate and the non mevalonate pathways. Plant Cell Rep. 2007;26:2129–36.
Weathers PJ, Elkholy S, Wobbe KK. Artemisinin: the biosynthetic pathway and its regulation in Artemisia annua, a terpenoid-rich species in vitro. Cell Dev Biol Plant. 2006;42:309–17.
Vergauwe A, Geldre EV, Inze D, Van Montagu M, Eeckhout VD. Factors influencing Agrobacterium tumefaciens-mediated transformation of Artemisia annua L. Plant Cell Rep. 1998;18:105–10.
Han JL, Liu BY, Ye HC, Wang H, Li ZQ, Li GF. Effects of overexpression of the endogenouse farnesyl diphosphate synthase on the artemisinin content in Artemisia annua L. J Integr Plant Biol. 2006;48:482–7.
Chen DH, Liu CJ, Ye HC, Li GF, Liu BY, Meng YL, et al. Ri-mediated transformation of Artemisia annua with a recombinant farnesyl diphosphate synthase gene for artemisinin production. Plant Cell, Tissue Organ Cult. 1999;57:157–62.
Chen D, Ye H, Li G. Expression of a chimeric farnesyl diphosphate synthase gene in Artemisia annua L. transgenic plants via Agrobacterium tumefaciens-mediated transformation. Plant Sci. 2000;155:179–85.
Kovalenko PG, Antonjuk VP, Maliutal SS. Secondary metabolites synthesis in transformed cells of Glycyrrhiza glabra and Potentilla alba as producents of radioprotective compounds. Bioorg Acta. 2004;1:13–22.
Weathers PJ, Bunk G, Mccoy MC. The effect of phytohormones on growth and artemisinin production in Artemisia annua hairy roots in vitro. Cell Dev Biol Plant. 2005;41:47–53.
Kim Y, Wyslouzil BE, Weathers PJ. A comparative study of mist and bubble column reactors in the in vitro production of artemisinin. Plant Cell Rep. 2001;20:451–5.
Kudakasseril GJ. Effect of sterol inhibitors on the incorporation of 14 C-isopentenyl pyrophosphate into artemisinin by a cell-free system from Artemisia annua tissue cultures and plants. Planta Med. 1987;53:280–4.
Arsenault PR, Vail DR, Wobbe KK, Weathers PJ. Effect of sugars on artemisinin production in Artemisia annua L. transcription and metabolite measurements. Molecules. 2010;15:2302–18.
Olofsson L, Anneli L, Peter EB. Trichome isolation with and without fixation using laser micro dissection and pressure catapulting followed by RNA amplification: expression of genes of terpene metabolism in apical and sub-apical trichome cells of Artemisia annua L. Plant Sci. 2011;183:9–13.
Schramek N, Wang H, Römisch-Margl W, Keil B, Radykewicz T, Winzenhörlein B, et al. Artemisinin biosynthesis in growing plants of Artemisia annua. A 13CO2 study. Phytochemistry. 2010;71:179–87.
Duke MV, Paul RN, Elsohly HN, Sturtz G, Duke SO. Localization of artemisinin and artemisitene in foliar tissues of glanded and glandless biotype of Artemisia annua L. Int J Plant Sci. 1994;155:365–72.
Liu B, Wang H, Du Z, Li G, Zi H. Metabolic engineering of artemisinin biosynthesis in Artemisia annua L. Plant Cell Rep. 2011;30:689–94.
Baldi A, Dixit VK. Yield enhancement strategies for artemisinin production by suspension culture of Artemisia annua. Bioresour Technol. 2008;99:4609–14.
Wang H, Ma C, Li Z, Ma L, Wang H, Ye H, et al. Effects of exogenous methyl jasmonate on artemisinin biosynthesis and secondary metabolism in Artemisia annua L. Industrial Crops Products. 2010;31:214–8.
Arsenault PR, Vail DR, Wobbe KK, Weathers PJ. Effect of sugars on artemisinin production in Artemisia annua L. transcription and metabolite measurements. Molecules. 2010;15:2302–18.
Duke SO, Vaughn KC, Edward M, Croom JR, Elsohly HN. Artemisinin, a constituent of annual wormwood (Artemisia annua), is a selective phytotoxin. Weed Sci. 1987;35:499–505.
Sambrook LF, Adams TR, Schell JS, Nester EW. Agrobacterium tumefaciens mediated transformation of Artemisia abscinthium stem explants by using kanamycin selection. Proc Natl Acad Sci. 1989;85:5536–40.
Authors’ contributions
BHK has performed all the experimental work and helped in data arrangement and wrote manuscript. JS has helped in artemisinin analysis and guidance in practical work. BM supervisor of the Project and reviewer. All the authors have read and approved the final manuscript.
Acknowledgements
This work is supported by the Higher education commission (HEC) Pakistan. The authors are very thankful to David Tepfer, Station d’Amélioration des Plantes, CNRS/INRA, Cedex, France for providing Agrobacterium strains. The authors are also very thankful to Guy Barker, Department of Life Sciences, for his help and guidance during this research work and Martin Davis from Department of Engineering, University of Warwick for help and cooperation in using Environmental scanning electron microscope.
Competing interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Kiani, B.H., Suberu, J. & Mirza, B. Cellular engineering of Artemisia annua and Artemisia dubia with the rol ABC genes for enhanced production of potent anti-malarial drug artemisinin. Malar J 15, 252 (2016). https://doi.org/10.1186/s12936-016-1312-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-016-1312-8