Abstract
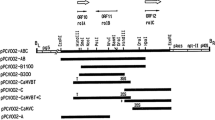
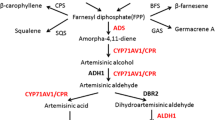
Artemisinin-based medicines are the most effective treatment for malaria. To date, wormwood plants (Artemisia annua L.) are the main source of artemisinin. Due to the limited nature of this source, considerable efforts are directed towards the development of methods of artemisinin production via heterologous expression systems. The goal of the study was to obtain tobacco transformed with genes of the artemisinin biosynthesis pathway and to analyze their transcription in a heterologous host. Tobacco plants were transformed with the artemisinin biosynthesis genes encoding amorpha-4,11-diene synthase, artemisinic aldehyde Δ11(13) reductase, amorpha-4,11-diene monooxygenase, and cytochrome P450 reductase from A. annua and yeast 3-hydroxy-3-methylglutaryl-coenzyme A reductase cloned in the pArtemC vector; farnesyl diphosphate synthase and aldehyde dehydrogenase were used to transform the plants as parts of vector p2356. Two transgenic lines with all target genes were obtained as a result of transformation with the pArtemC and p2356 vectors. After double transformation with these vectors, six transgenic lines were obtained. Five genes of artemisinin biosynthesis and two genes of biosynthesis of its precursors were successfully transferred into the genome of transgenic tobacco lines as a result of the cotransformation with the abovementioned vectors. Thus, the entire artemisinin biosynthesis pathway was first reconstructed in heterologous plants: the transcription of the artemisinin biosynthesis genes in the tobacco plants was shown via RT-PCR. The results will be used in further research on expression systems for the production of artemisinin and other nonprotein substances in heterologous host plants.




Similar content being viewed by others
REFERENCES
Ho, W., Peh, H., Chan, T., et al., Artemisinins: pharmacological actions beyond anti-malarial, Pharmacol. Ther., 2014, vol. 142, no. 1, pp. 126–139. https://doi.org/10.1016/j.pharmthera.2013.12.001
Brown, G., The biosynthesis of artemisinin (Qinghaosu) and the phytochemistry of Artemisia annua L. (Qinghao), Molecules, 2010, vol. 15, pp. 7603–7698. https://doi.org/10.3390/molecules15117603
Ikram, N.K.B.K. and Simonsen, H.T., A review of biotechnological artemisinin production in plants, Front. Plant Sci., 2017, vol. 8, p. 1966. https://doi.org/10.3389/fpls.2017.01966
Tu, Y., Metabolic engineering of Artemisia annua L., chapter 8 in From Artemisia annua L. to Artemisinins. The Discovery and Development of Artemisinins and Antimalarial Agents, Chemical Industry Press, 2017, pp. 163–208. https://doi.org/10.1016/B978-0-12-811655-5.00008-8
Wen, W. and Yu, R., Artemisinin biosynthesis and its regulatory enzymes: progress and perspective, Pharmacogn. Rev., 2011, vol. 5, pp. 189–194. https://doi.org/10.4103/0973-7847.91118
Teoh, K., Polichuk, D., Reed, D., et al., Artemisia annua L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin, FEBS Lett., 2006, vol. 580, no. 5, pp. 1411–1416. https://doi.org/10.1016/j.febslet.2006.01.065
Teoh, K., Polichuk, D., Reed, D., et al., Molecular cloning of an aldehyde dehydrogenase implicated in artemisinin biosynthesis in Artemisia annua, Botany, 2009, vol. 87, no. 6, pp. 635–642. https://doi.org/10.1139/B09-032
Farhi, M., Marhevka, E., Ben-Ari, J., et al., Generation of the potent anti-malarial drug artemisinin in tobacco, Nat. Biotechnol., 2011, vol. 29, no. 12, pp. 1072–1074. https://doi.org/10.1038/nbt.2054
Ikram, N., Beyraghdar Kashkooli A., Peramuna A., et al. Stable production of the antimalarial drug artemisinin in the moss Physcomitrella patens, Front. Bioeng. Biotechnol., 2017, vol. 5, p. 47. https://doi.org/10.3389/fbioe.2017.00047
Malhotra, K., Subramaniyan, M., Rawat, K., et al., Compartmentalized metabolic engineering for artemisinin biosynthesis and effective malaria treatment by oral delivery of plant cells, Mol. Plant., 2016, vol. 9, no. 11, pp. 1464– 1477. https://doi.org/10.1016/j.molp.2016.09.013
Wang, B., Kashkooli, A., Sallets, A., et al., Transient production of artemisinin in Nicotiana benthamiana is boosted by a specific lipid transfer protein from A. annua, Metab. Eng., 2016, vol. 38, pp. 159–169. https://doi.org/10.1016/j.ymben.2016.07.004
Han, J., Liu, B., Ye, H., et al., Effects of overexpression of the endogenous farnesyl diphosphate synthase on the artemisinin content in Artemisia annua L., J. Integr. Plant Biol., 2006, vol. 48, no. 4, pp. 482–487. https://doi.org/10.1111/j.1744-7909.2006.00208.x
Banyai, W., Kirdmanee, C., Mii, M., et al., Overexpression of farnesyl pyrophosphate synthase (FPS) gene affected artemisinin content and growth of Artemisia annua L., Plant Cell Tissue Organ Cult., 2010, vol. 103, no. 2, pp. 255–265. https://doi.org/10.1007/s11240-010-9775-8
Tang, K.X., Chen, Y.F., Shen, Q., et al., Overexpression aldh1 gene increased artemisinin content in Artemisia annua L., CN Patent CN201210014242.7, Shanghai: Jiao Tong University, China, August 22, 2012.
Collier, R., Thomson, J.G., and Thilmony, R., A versatile and robust Agrobacterium-based gene stacking system generates high-quality transgenic Arabidopsis plants, Plant J., 2018, vol. 95, no. 4, pp. 573–583. https://doi.org/10.1111/tpj.13992
Untergasser, A., Bijl, G.J., Liu, W., et al., One-step Agrobacterium mediated transformation of eight genes essential for Rhizobium symbiotic signaling using the novel binary vector system pHUGE, PLoS One, 2012, vol. 7, no. 10. e47885. https://doi.org/10.1371/journal.pone.0047885
Fujisawa, M., Takita, E., Harada, H., et al., Pathway engineering of Brassica napus seeds using multiple key enzyme genes involved in ketocarotenoid formation, J. Exp. Bot., 2009, vol. 60, no. 4, pp. 1319–1332. https://doi.org/10.1093/jxb/erp006
Ward, J., Xu, J., Lamb, J., et al., The use of Agrobacterium tumefaciens to deliver ten genes on a single vector to corn, in The 9th International Plant Molecular Biology (IPMB) Congress, 2009, 2009, poster #1364.
Chung, S.M., Frankman, E.L., and Tzfira, T., A versatile vector system for multiple gene expression in plants, Trends Plant Sci., 2005, vol. 10, no. 8, pp. 357–361. https://doi.org/10.1016/j.tplants.2005.06.001
Horsch, R.B., Fry, J.E., Hofmann, N.L., et al., A simple and general method for transferring genes into plants, Science, 1985, vol. 227, pp. 1229–1231.
Dellaporta, S.L., Wood, J., and Hicks, J.B., A plant DNA minipreparation version II, Plant Mol. Biol. Rep., 1983, vol. 1, pp. 19–21.
Tzfira, T., Lacroix, B., and Citovsky, V., Nuclear import of Agrobacterium T-DNA, in Madame Curie Bioscience Database [Internet], Austin (TX): Landes Bioscience, 2000–2013.
Gelvin, S.B., Integration of Agrobacterium T-DNA into the plant genome, Annu. Rev. Genet., 2017, vol. 51, pp. 195–217. https://doi.org/10.1146/annurev-genet-120215-035320
ACKNOWLEDGMENTS
The work was carried out using at unique scientific facility Fitotron, reg. no. 2-2.9.
Funding
The work was supported by the Russian Science Foundation, grant no. 19-14-00190.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflicts of interest.
This article does not contain any studies involving animals performed by any of the authors.
This article does not contain any studies involving human participants performed by any of the authors.
Additional information
Translated by I. Gordon
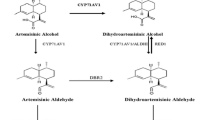
Abbreviations: AD—amorpha-4,11-diene; ADS—amorpha-4,11-diene synthase; ALDH1—aldehyde dehydrogenase; BAP—6‑benzylaminopurine; CaMV—cauliflower mosaic virus; CPR—cytochrome P450 reductase; CYP71AV1—amorpha-4,11-diene monooxygenase; DBR—2Δ11(13) reductase; FPP—farnesyl diphosphate; FDPS—farnesyl diphosphate synthase; IAA—Δ-indolylacetic acid; IPP—isopentyl diphosphate; MS medium—Murashige–Skoog medium; MVA—mevalonate; OD600—optical density at a wavelength of 600 nm; Rubisco-ribulose-1,5-bisphosphate carboxylase/oxygenase; RT-PCR—polymerase chain reaction with reverse transcription; tHMGR—3-hydroxy-3‑methylglutaryl coenzyme A reductase.
Rights and permissions
About this article
Cite this article
Firsov, A.P., Mitiushkina, T.Y., Pushin, A.S. et al. Agrobacterial Transformation of Tobacco with a Genetic Module of the Biosynthesis of the Antimalarial Agent Artemisinin. Appl Biochem Microbiol 57, 808–817 (2021). https://doi.org/10.1134/S0003683821070024
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0003683821070024