Abstract
Background
The circular RNA (circRNA) plays a vital role in the pathogenesis of tumors as a competitive endogenous RNA (ceRNA). Given the high aggressiveness and fatality rate of glioblastoma (GBM) as well as poor prognosis, it is necessary to construct a circRNA-related ceRNA network for further studies on the mechanism of GBM and identify possible biomarkers as well as therapeutic drugs.
Methods
Three datasets from the gene expression omnibus (GEO) database were downloaded to distinguish differential circRNAs, microRNAs, and messenger RNAs respectively in GBM. With the help of GEPIA2, circBank, CSCD, TargetScan, miRDB, and miRTarBase databases, we established a circRNAs-related ceRNA network in GBM. Functional enrichments were employed to profile the most relevant mRNAs to indirectly clarify the mechanisms of the ceRNA network. Based on the expression profile data and survival information of GBM patients from the GEO and the cancer genome atlas (TCGA) databases, we performed survival analysis to select prognostic mRNAs and constructed a novel circRNA-miRNA-mRNA central regulatory subnetwork. The DGIdb database was used to find potential drug–gene interactions.
Results
The datasets obtained from the GEO and TCGA databases were analyzed, and 504 differentially expressed mRNAs (DEmRNAs), 71 differentially expressed microRNAs (DEmiRNAs), and 270 differentially expressed circRNAs (DEcircRNAs) were screened out. The novel ceRNA regulatory network included 22 circRNAs, 11 miRNAs, and 15 mRNAs. FZD1 and KLF10 were significantly correlated with the overall survival rate of patients with GBM (P < 0.05). The final survival subnetwork contained six circRNAs, two miRNAs, and two mRNAs. Two small-molecule compounds and one antibody could be used as therapeutic drugs for GBM. Interestingly, the Wnt signaling pathway appeared in both KEGG and GO functional terms.
Conclusions
Results of this study demonstrate that FZD1 and KLF10 may exert regulatory functions in GBM, and the ceRNA-mediated network could be a therapeutic strategy for GBM.
Similar content being viewed by others
Background
Glioblastoma is the most frequent primary malignant brain tumor in adults and is often rapidly fatal [1]. The associated complications impose a substantial economic burden and medical strain around the world [2]. Despite advances in current treatments, their impact on GBM remains limited, possibly due to an insufficient understanding of its pathogenesis. Hence, more studies must be conducted to investigate the mechanisms of GBM and to identify novel biomarkers for predicting prognosis and treatment outcomes.
Using high-throughput sequencing techniques, several noncoding RNA (ncRNAs) were found to be correlated with dysregulated gene expression and biological process imbalances in GBM [3].
CircRNAs are unaffected by exonucleases (more stable than linear RNAs) and typically exhibit tissue or developmental stage-specific expression, implying their biological function in various diseases, including GBM [4]. In line with this, the competing endogenous RNA (ceRNA) hypothesis mentions that circRNAs act as miRNA sponges in cells to bind to them and affect the corresponding target genes [5]. For instance, circNT5E controls various pathological processes such as cell proliferation, metastasis, and invasion, and is rich in miR-422a binding sites. CircNT5E can inhibit miR-422a activity by binding, showing tumor suppressor-like characteristics in GBM [6]. Besides, the functions of circRNAs (including circCCDC66, circSMARCA5, circMTO1) have been gradually discovered, especially their ability to bind miRNAs in cervical cancer, hepatocellular carcinoma, and glioma pathology [7,8,9,10]. Therefore, we hypothesize that the expression of miRNA is negatively correlated with the expression of circRNA and mRNA. The importance of ceRNA in tumorigenesis is self-evident, but the roles of the circRNA–mRNA–miRNA ceRNA network in GBM remain unclear.
This research involved the comprehensive analysis of multiple databases. Furthermore, this research analyzed the effect of differentially expressed genes (DEGs) on GBM overall survival, constructed a prognosis-related subnetwork to strengthen the understanding of the pathogenesis of GBM, and proposed several drugs that might be potential therapeutic agents for GBM.
Materials and methods
Data obtained
We downloaded expression profiles of the GSE165926, GSE103229, and GSE90886 datasets from the GEO database web resource (https://www.ncbi.nlm.nih.gov/geo/). The GSE165926 dataset contained circRNA data from 4 normal brains and 12 GBM tissues [11]. The GSE103229 dataset, which comprised microRNA expression profiling, included 5 normal brain tissues and 5 GBM tissues. The GSE90886 dataset included mRNA expression profiles of 9 normal tissues and 9 GBM tissues [12]. To verify the preliminary findings, the serial expression profiles of glioblastoma were downloaded from the TCGA database (http://cancergenome.nih.gov/), including data from 5 non-tumor brains and 169 GBM samples for survival analysis and collection of clinical information on cases.
Identification of differentially expressed genes (DEGs)
The annotation documents of the corresponding platforms were used for gene expression profiling. The average value of gene expression was used when one gene corresponded to multiple probes. The “LIMMA” package in R software (version 4.1.1) was employed to identify differentially expressed miRNAs, circRNAs, and mRNAs between glioblastomas and non-tumor brain tissues in both GEO datasets and TCGA datasets, with thresholds respectively set at |log2 fold change (FC)|≥ 2.0, 2.0, 1.0; P < 0.05 [13]. Next, the differentially expressed genes (DEGs) from the GEO and TCGA datasets were divided into upregulated and downregulated groups for further analysis. Venn diagrams were drawn using the “Venn” package of R software (version 4.1.1).
ceRNA network establishment
Firstly, we converted the names of DEcircRNAs to mature circRNA names using the circBASE database web source (http://www.circbase.org/). Then the CSCD database (web source: http://gb.whu.edu.cn/CSCD/) was employed to find the miRNAs response elements (MREs) by entering circRNAs names. Only the interactions supported by solid evidence (reporter assays, western blots, and qPCR) were included in the prediction to enhance the reliability of the results. To further investigate the role of mRNAs in GBM, we used the TargetScan (http://www.targetscan.org/), miRTarBase (http://mirtarbase.cuhk.edu.cn/php/index.php), and miRDB (https://www.mirbase.org/) databases to find miRNAs target genes [14]. We compared the target genes and DEmRNAs, retaining only data for intersecting mRNAs and corresponding miRNAs for further study. According to the prediction and evaluation above, a visualized circRNAs-miRNAs-mRNAs network was constructed using Cytoscape software (version 3.8.2,
http://www.cytoscape.org/). Statistical significance was only determined for terms with a P < 0.05.
Functional enrichment analysis
ClusterProfiler is an R package that provides three methods, enrichGO, enrichKEGG, and groupGO, for enrichment analysis and classification [15]. We used it to perform the Kyoto Encyclopedia of Genes and Genome (KEGG) and Gene Ontology (GO) pathway enrichment analyses of DEmRNAs in the circRNAs-miRNAs-mRNAs network. P < 0.05 was set as the criterion for statistical significance, and the terms were visualized via the “ggplot2” package in R.
Construction of a protein–protein interaction (PPI) network
A PPI network was established by the Search Tool for the Retrieval of Interacting Gene (STRING, https://string-db.org/), an online database of billions of interactions for thousands of organisms [16] and visualized via Cytoscape software (version 3.8.2). The list of DEmRNAs was first mapped to the STRING locus, and their interactions were evaluated. In addition, the genes were selected based on the PPIs comprehensive score > 0.9 and the degree of close correlation with other genes adjusted to ≥ 10 [16]. The Molecular Complex Assay (MCODE) based on a weighted algorithm was then used to identify highly interacting gene modules in the PPI network, and the standard parameters were set by default, except k-core = 7 [17].
Survival analysis and prognostic network construction
To determine critical genes associated with the survival of GBM patients, the prognostic values of the DEmRNAs were estimated with the clinical information acquired from TCGA. The “survival” package in R software was employed to find associations between mRNAs and survival status data [18]. FDR < 0.05 was statistically significant. The association of FZD1 (Frizzled class receptor 1) and KLF10 (Krüppel-like factor 10) with immune infiltration in GBM was analyzed in an online database: TIMER (web source: https://cistrome.shinyapps.io/timer/), which can investigate immune cell infiltration levels using data from TCGA. We used Cytoscape software (version 3.8.3) to construct the prognostic circRNA-miRNA-mRNA ceRNA subnetwork.
Drug–gene interaction
Through the interaction, DEmRNAs were paired with existing compounds to discover potential therapeutic agents for GBM. The drug–gene interaction database (DGIdb, web source: https://dgidb.org/) provides an interface for searching gene lists based on drug–gene interaction profiles and potentially "druggable" genes [19]. DEmRNAs were uploaded into DGIdb to match with the existing drugs.
Meta-analysis
To validate our findings, the eligible published studies were searched by the following databases: Chinese CNKI, Cochrane Library, Web of Science, and PubMed. Citations from the inclusive studies were also selected for additional relevant studies. We used “FZD1” or “fzd1” or “Frizzled class receptor 1” AND “neoplasms” or “cancer” or “tumor” or “carcinoma”, “KLF10” or “klf10” or “Krüppel-like factor 10” AND “neoplasms” or “cancer” or “tumor” or “carcinoma” as the search terms for appropriate identification. The exclusion and inclusion criteria are shown in Additional file 1: Table 1. We extracted the following information: 1st author, country, date of publication, patients’ number, cancer type, sample assay approach, threshold value of FZD1 and KLF10 expression level, 95% CI and HR of overall survival (OR). We recorded 95% CI and HR directly if they were available in the studies. If not, we analyzed and abstracted the data from Kaplan–Meier (K–M) curves of OS based on the approach described by a previous research [20]. Two researchers evaluated the data independently, a third investigator decided whether to incorporate the study. The Newcastle–Ottawa Scale (NOS) was used to evaluate the quality of the enrolled studies [21].
Statistics
Statistical analyses were performed with the R software (version 4.1.1) and the above packages. Meta-analysis was conducted using the SPSS software. Chi-square-based I2 tests and Cochran’s Q were conducted to test for heterogeneity. I2 < 50%, P > 0.10 indicates no significant heterogeneity across studies. Subgroup analyses were applied to investigate the sources of heterogeneity. The 95% CI and HR in each study were integrated and plotted as forest plots to assess the impact of FZD1 and KLF10 expressions on OS in cancers. Sensitivity analysis was used to examine the reliability of the results. Two-tailed P values < 0.05 were considered statistically significant.
Results
DEGs detection
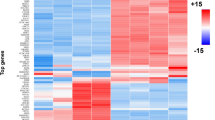
Figure 1 displays the basic flow chart of this study. Firstly, 270 DEcircRNAs, including 183 down-regulated genes and 87 up-regulated genes, were identified from normal controls and GBM samples in the GSE165926 dataset by the “limma” package of R software, by setting the cutoff value as |log2 fold change (FC)|≥ 2, P < 0.05; Similarly, 71 DEmiRNAs were screened in the GSE103229 dataset, including 34 down-regulated and 37 up-regulated genes; the criterion was set as |log2 FC|≥ 2, P < 0.05. Meanwhile, the GBM samples and normal control group in the GSE90886 dataset were analyzed, and 504 DEmRNAs were obtained, including 242 down-regulated genes and 262 up-regulated genes, with thresholds set at |log2 FC|≥ 1, P < 0.05. We showed the heatmaps of some DEGs and several circRNAs in Fig. 2A–C. The structures of several DEcircRNAs are displayed in Fig. 2D.
Expression profile heatmaps for DEGs were screened from three datasets in the ceRNA network. A represents the heatmap of circRNAs, B represents the miRNAs' heatmap, and C represents the mRNAs' heatmap. D Structure diagrams of circRNAs; the different colors, shapes of the outer and inner loops represent the different exons and locations of MRE, RBP, and OR
Construction of the ceRNA network
To further ensure the reliability of differentially expressed RNAs and the ceRNA network in GBM, we only retained the overlapping parts from the three datasets, we selected circRNAs-miRNAs interaction pairs and miRNAs-mRNAs interaction pairs from TargetScan, miRDB, and miRTarBase databases to construct a circRNAs-miRNAs-mRNAs ceRNA network. Results showed that 44 of 71DEmiRNAs in the GSE103229 dataset were the same as the integrated miRNAs in the GSE165926 dataset, and 48 of 504 DEmRNAs in the GSE90886 dataset corresponded with the miRNA-targeted genes likewise (Fig. 3). A total of 11 DEcircRNAs and 15 DEmRNAs constituted 17 interacting pairs. In addition, 22 DEcircRNAs and 11 DEmiRNAs formed 24 interaction pairs. Afterwards, 22 DEcircRNAs, 15 DEmRNAs, and 11 DEmiRNAs were used to establish a ceRNA network using Cytoscape software (version 3.8.2) for visualization (Fig. 4). We also drew the boxplots of DEcircRNAs, DEmiRNAs, and DEmRNAs in the ceRNA network, respectively (Fig. 5A–C).
The ceRNA network about circRNAs-miRNAs-mRNAs. Light red triangular nodes represent the downregulated miRNAs, dark red triangular nodes represent the upregulated miRNAs; Light green diamonds represent the downregulated; dark green diamonds represent the upregulated circRNAs; Light blue elliptical nodes represent the downregulated, dark blue elliptical nodes represent the upregulated mRNAs
Function and pathway enrichment analysis
We performed the KEGG pathway and GO functional enrichment analysis using DEmRNAs in the ceRNA network. GO results revealed that DEmRNAs were more related to the ossification (BP), Wnt signalosome (CC), and phosphoric diester hydrolase activity (MF) (Fig. 6A–C; Table 1), respectively. KEGG analysis revealed 19 statistically significant enriched pathways. The top-3 most enriched pathways were: Melanogenesis, Cushing syndrome, Wnt signaling pathway (Fig. 6D; Table 2).
PPI network and module analysis
We investigated the relationship between DEmRNAs in the network through the online tool STRING database. Then we use the plugin MCODE to select a more densely connected module with 30 edges, 17 nodes and a score of > 0.900(the highest confidence level) in this PPI network. (Fig. 7).
DEmRNAs associated with the overall survival of GBM patients
To investigate the prognostic ability of DEmRNAs in GBM, 15 DEmRNAs in the circRNA-associated ceRNA networks were analyzed using the "survival" package of the R software (version 4.1.1). The results indicated that 2 out of 15 DEmRNAs were significantly correlated with the overall survival in GBM (FDR < 0.05). The two DEmRNAs were FZD1 and KLF10 and were inversely correlated with overall survival, as is shown in Fig. 8A, B. To better understand the roles of two hub genes in the GBM immune microenvironment, we analyzed the association between FZD1, KLF10, and several common immune cell types in the TIMER database. As shown in Fig. 8C, D, FZD1 was positively correlated with the infiltrations of CD4 + T cells, macrophages, neutrophils, and dendritic cells in GBM. Strong positive correlations existed between KLF10 expression and the infiltrations of neutrophils and dendritic cells in GBM. Finally, the related ceRNA subnetwork was also constructed using the Cytoscape software (version 3.8.2), as shown in Fig. 9.
A, B Association between overall survival and mRNAs in this network analyzed via K–M survival curves. Two mRNAs were significantly correlated with the overall survival of GBM patients (FDR < 0.05), they were FZD1 and KLF10. C, D The correlation between FZD1, KLF10 and immune infiltration of GBM was studied using the TIMER database
Drug–gene interaction
Drug–gene interactions analysis was performed on the 15 mRNAs in this circRNA-associated ceRNA network. According to the results, four drugs interacted with gene FZD1 (frizzled class receptor (1), six drugs significantly interacted with ENPP2 (ectonucleotide pyrophosphatase /phosphodiesterase (2). At the same time, DYRK3 (dual specificity tyrosine-phosphorylation-regulated kinase (3) were associated with 11 different drugs. Of the two drugs, only three were found to have antineoplastic activities in GBM therapy and targeted FZD1 and ENPP2 genes, respectively. The 2D structural diagrams of these compounds are shown in Additional file 2: Figure S1.
The relationship between FZD1 and KLF10 expression and prognosis
We performed the meta-analysis to initially validate the prognosis ability of FZD1 and KLF10. Based on the above-mentioned criteria, six FZD1-related articles [22,23,24,25,26,27] including 972 patients were finally included and five KLF10-associated articles [28,29,30,31,32], consisting of 665 patients, were ultimately enrolled. Specific information on these studies is presented in Additional file 1: Tables 2 and 3. The results revealed that high expression of FZD1 was significantly associated with poor OS (HR 2.55; 95% CI 1.95–3.34; P < 0.00001) (Additional file 2: Fig. 2A). The heterogeneity of the data was not significant (I2 = 39%, P = 0.15), therefore the fixed-effect model was applied. As presented in Additional file 2: Fig. 2B, high KLF10 expression was significantly correlated with an inferior OS (HR 1.41; 95% CI 1.02–1.97; P = 0.04). There was no heterogeneity (I2 = 16%, P = 0.32), so a fix-effects model was used. Subsequent subgroup analyses were conducted by analyzing the record method, detecting approach, and cancer type to stratify patients. The results are shown in Additional file 1: Tables S4 and S5. And no significant bias was found by performing the funnel plot analysis (Additional file 2: Fig. 3A, B).
Discussion
Glioblastoma (GBM) is one of the deadliest cancers in humans [33]. Treatment guidelines recommend surgical resection for newly diagnosed glioblastoma, followed by adjuvant radiation therapy and adjuvant chemotherapy [34]. Despite an aggressive treatment approach for GBM patients, median survival is generally less than one year from the date of diagnosis, and most patients die within two years, even in the most favorable circumstances [35]. Differential genes have been reported to play a key role in tumor pathology and thus may be potential targets for tumor therapy [36, 37]. This study investigated the pathological mechanism of GBM to determine and identify potential therapeutic agents for GBM. Previous studies have shown that circRNAs, miRNAs, and mRNAs play a significant role in the pathological formation of tumor processes [37,38,39], but little attention has been paid to the overall regulatory role of these genes in tumors, including GBM.
In this research, we mined three datasets from the GEO database to identify differentially expressed circRNAs, miRNAs, and mRNAs between normal brains and GBM tissue samples. The intersecting miRNAs and mRNAs were selected from these three datasets using bioinformatics techniques, and a circRNAs-miRNAs-mRNAs ceRNA network was established. The functions and enrichment pathways of mRNAs in this network were further explored by the GO and KEGG analyses. We then downloaded the clinical data of GBM from the TCGA database, analyzed the clinical information of relevant genes in this network, and identified 2 mRNAs, 6 circRNAs, and 2 miRNAs associated with the overall survival of GBM. Ultimately, the online drug database DGIdb was used to analyze the DEmRNAs in this network and screen three compounds that could be potential drugs for GBM.
GO analysis showed that mRNAs were enriched in some major regions, such as ossification, Wnt signalosome, and phosphoric diester hydrolase activity. And the KEGG results indicated that the top-3 most enriched pathways were Melanogenesis, Cushing syndrome, and Wnt signaling pathway, some of which have been reported in previous research regarding cancer. Melanogenesis plays an critical regulatory role in the differentiation, proliferation, apoptosis, and migration of GBM and melanoma [40]. Paraneoplastic Cushing's syndrome, caused by ectopic adrenocorticotropic hormone (ACTH) secretion, has been reported to be associated with neuroendocrine prostate cancer, small-cell lung cancer, and a variety of tumors [41, 42].
Interestingly, we found that the "Wnt signaling pathway" appeared in both GO and KEGG enrichment analyses. The WNT (Wingless/Integral) signaling pathway is involved in different stages of GBM. It has been recognized as a marker of a therapeutic challenge due to its critical function in normal tissue homeostasis [43]. Wnt signaling acts as an essential regulator of critical cellular events in the developing brain. More specifically, it is essential for self-renewal, differentiation, proliferation, and migration in neurons, astrocytes, oligodendrocytes, and so on [44]. Numerous studies have shown that excessive activation of Wnt signaling was associated with the malignant transformation and development of brain tumors [45,46,47]. Diclofenac and celecoxib inhibit tumor cell proliferation in a variety of human malignancies and are potential therapeutic drugs targeting glioblastoma cells that work by inhibiting activation of Wnt/β-catenin/Tcf signaling [48].
This constructed ceRNA network consisted of the core DEGs after being intersected from three datasets. We believe that this network contributes to further understanding of GBM pathogenesis at the genetic level. Some of these interactions have been demonstrated in previous reports. For instance, MiR-144, a tumor suppressor, inhibits cell viability, invasion and cell migration by downregulating CEP55 and induces apoptosis and cell cycle arrest in breast cancer [49]. This discovery provided new prognostic biomarkers and therapeutic targets for the development of breast cancer diagnosis and treatment. Also, MiR-494 inhibits the expression of the oncogene HOXA10 and thereby reduces the proliferation of oral cancer tissue cells [50]. And overexpression of circRNA_102171 has been reported to promote the occurrence and progression of papillary thyroid carcinoma (PTC) by activating the Wnt/β-catenin pathway in a CTNNBIP1-dependent manner [51]. In addition, several mRNAs in this ceRNA network have been found to have regulatory roles in GBM, such as VSNL1, CEP55, HOXA10, ENPP2 [52,53,54,55], and so on. We believe that with the deepening of GBM basic and clinical trials, more and more gene targets and corresponding therapeutic drugs will be found in this ceRNA network.
To confirm the credibility of our analysis, we again built a PPI network with DEmRNA, where 12 mRNAs in the ceRNA network appeared in the most important modules. The 12mRNAs were: NIPAL3, CEP55, VSNL1, FZD1, HOXA10, PLCB1, SOX11, ENPP2, ELAVL3, DDX-21, KLF10, DYRK3, some of which have been found to play an important role in the pathological process of GBM. Clinical information, including overall survival, downloaded from the TCGA database was used to analyze the prognostic ability of genes in the circRNAs-associated ceRNA network. Six of 22 circRNAs, two of 11 miRNAs, and two of 15 mRNAs were associated with the patient's overall survival rate. These genes were miR-224-3p, miR-4762-5p, circ_0016956, circ_0006841, circ_0006635, circ_0056281, circ_0005223, circ_0005777, FZD1, KLF10. Among them, we focused on the downregulated mRNAs (FZD1 and KLF10) significantly correlated with overall survival in GBM.
It has been reported in many cancer tissues that overexpression of FZD1 can lead to tumor progression and drug resistance [25]. FZD1 is the first member of the Frizzled gene family that translates seven-transmembrane proteins, which are receptors for the Wnt signaling pathway [56]. FZD1 could also regulate the cellular response in physiological and pathological microenvironmental conditions [57]. Previous research found that the gradient effect of FZD1 expression in the tumor microenvironment may regulate colon cancer progression and spread, providing a new therapeutic target for colon cancer patients [58]. FZD1 appears to mediate drug resistance by modulating the Wnt/β-catenin pathway in clear cell renal cell carcinoma [25], neuroblastoma [59], pancreatic ductal adenocarcinoma [27], ovarian cancer [60], and breast cancer [61]. KLF10 (Krüppel-like factor 10) has been established in several studies for its role as a tumor suppressor in cancer [62]. KLF10 has the potential to be a marker of various diseases, including cardiac hypertrophy [62], diabetes [63], osteoporosis [64], immune system diseases, and colitis [65]. KLF10 plays a significant role in many conditions and biological processes, including tumorigenesis. The expression of KLF10 was negatively correlated with the progressive worsening of pancreatic cancer; therefore, KLF10 may be used as a predictor of pancreatic cancer staging [28]. Moreover, KLF10 expression can also serve as an independent prognostic marker in oral cancer patients, especially those in early-stage [30]. KLF10 inhibits β-catenin nuclear transformation and Wnt signaling pathways in vivo, supporting its potential therapeutic target of multiple myeloma [66]. Research by Marrero, D et al. found that changes in KLF10 and mRNA expression and variation in PSG copy number may be new molecular markers of cervical cancer [67]. The results of this study further demonstrate the potential of hub genes involved in the network in assessing the prognosis of GBM.
Glioblastoma is one of the most drug-resistant malignancies and often recurs after chemotherapy. Therefore, there is some need to explore new compounds or drugs to achieve the desired therapeutic effect. Three drugs (Niclosamide, Vantictumab against FZD1; Ziritaxestat against ENPP2) were identified as potential candidates with antitumor activity that may play a role in the treatment of GBM.
Vantictumab is a fully-humanized monoclonal antibody that binds to the frizzled (FZD) receptor and inhibits the classical WNT signaling pathway [68]. A combination of vantictumab and paclitaxel has shown promising clinical activity in patients with metastatic or locally advanced breast cancer who have received 2 prior routes of chemotherapy [69]. In addition, in the study of limiting the dilution of pancreatic transplantation, vantictumab could reduce the frequency of activity of initiating tumor cells in vitro, further supporting its effect on tumor stem cells [69].
Niclosamide has significant inhibitory, cytotoxic, and anti-migratory effects. It can reduce the frequency of multipotent/self-renewing cells in vitro and the malignant potential of primary human glioblastoma cells in vivo [70]. Therefore, niclosamide, an anthelmintic drug used to treat tapeworm infections, has recently renewed interest in its use as a treatment for diseases such as colon cancer and glioblastoma. A variety of studies have uncovered that niclosamide had effects on cell inhibition, cytotoxicity, and anti-migration and inhibited intracellular NOTCH-, mTOR-, WNT/CTNNB1-, and NF-kB signal transduction [70,71,72,73]. Among them, Oh et al. discovered that the combination of temozolomide and niclosamide could effectively reduce the metastasis and aggressiveness of GBM tumorspheres [72]. The last drug, ziritaxestat, is in phase 3 clinical trials for the treatment of idiopathic pulmonary fibrosis (IPF) but has been reported to act as an autotaxin inhibitor for cancer treatment when combined in different ways with other small compounds [74].
As multiple TCGA and GEO datasets were included, tumor samples were not evaluated for GBM phenotypes, which may have influenced the expression profiles and prognosis of GBM patients. In addition, all survival analyses were based on GBM samples only and were performed using online databases with an automatic assessment of confounding factors. And the primary screening measure in this study was prognostic value, which may have resulted in the omission of potentially valuable information. In the part of meta-analysis, most of the included studies were from China and there may be potential publication bias. Currently, drugs and genes, including the constructed ceRNA network we have identified, rely heavily on bioinformatics tools and databases. Their accuracy still needs to be validated by basic experiments and clinical trials. Therefore, in this study, we adopted and integrated multiple databases and methods for all predictions to improve the fidelity of the data.
Conclusions
The role of ceRNAs in the progression of GBM has been reported, but only a few studies have focused on the part of ceRNAs in the prognosis of GBM patients. Furthermore, the majority of previous studies have constructed ceRNA networks based on sequential patterns of lncRNA–miRNA–mRNA in GBM. This study was one of the few to establish a GBM-related ceRNA network via mRNA–miRNA-circRNA sequential patterns. In conclusion, the findings of this study may provide new insights and potential therapeutic strategies for glioblastoma.
Availability of data and materials
The author confirms that data from the public database for this study have explained the source of the data in detail in the manuscript. The data can be found at the GEO data repository (https://www.ncbi.nlm.nih.gov/geo/) and TCGA data repository (http://cancergenome.nih.gov/), including the accession numbers: GSE165926, GSE103229, and GSE90886. And we have provided the related scripts used in this study and information of DEGs, including DEmRNAs, DEmiRNAs, and DEcircRNAs (Additional files 3, 4, 5, 6, 7).
Abbreviations
- DEGs:
-
Differentially expressed genes
- circRNAs:
-
Circular RNAs
- miRNAs:
-
Micro RNAs
- mRNAs:
-
Massager RNAs
- ceRNA:
-
Competitive endogenous RNA
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genome
- K–M:
-
Kaplan–Meier
- CI:
-
Confidence interval
- HR:
-
Hazard ratio
References
Ou A, Yung WKA, Majd N. Molecular mechanisms of treatment resistance in glioblastoma. Int J Mol Sci. 2021;22(1):351.
Wang G-M, Cioffi G, Patil N, Waite KA, Lanese R, Ostrom QT, et al. Importance of the intersection of age and sex to understand variation in incidence and survival for primary malignant gliomas. Neuro Oncol. 2022;24(2):302–10.
Zeng T, Li L, Zhou Y, Gao L. Exploring long noncoding RNAs in glioblastoma: regulatory mechanisms and clinical potentials. Int J Genom. 2018;2018:1–13.
Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–61.
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–8.
Wang R, Zhang S, Chen X, Li N, Li J, Jia R, et al. CircNT5E acts as a sponge of miR-422a to promote glioblastoma tumorigenesis. Can Res. 2018;78(17):4812–25.
Hsiao K-Y, Lin Y-C, Gupta SK, Chang N, Yen L, Sun HS, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Can Res. 2017;77(9):2339–50.
Han D, Li J, Wang H, Su X, Hou J, Gu Y, et al. Circular RNA circMTO1 acts as the sponge of MicroRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–64.
Zheng J, Liu X, Xue Y, Gong W, Ma J, Xi Z, et al. TTBK2 circular RNA promotes glioma malignancy by regulating miR-217/HNF1 beta/Derlin-1 pathway. J Hematol Oncol. 2017;10:351.
Yu J, Xu Q-G, Wang Z-G, Yang Y, Zhang L, Ma J-Z, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68(6):1214–27.
Li Y, Chen J, Chen Z, Xu X, Weng J, Zhang Y, et al. CircGLIS3 promotes high-grade glioma invasion via modulating Ezrin phosphorylation. Front Cell Dev Biol. 2021;9:663207.
Long H, Liang C, Zhang Xa, Fang L, Wang G, Qi S, et al. Prediction and analysis of key genes in glioblastoma based on bioinformatics. Biomed Res Int. 2017;2017.
Xiong D-D, Dang Y-W, Lin P, Wen D-Y, He R-Q, Luo D-Z, et al. A circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med. 2018;16:1–21.
Liu Y, Zhu J, Ma X, Han S, Xiao D, Jia Y, et al. ceRNA network construction and comparison of gastric cancer with or without Helicobacter pylori infection. J Cell Physiol. 2019;234(5):7128–40.
Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics J Integr Biol. 2012;16(5):284–7.
Zhou Z, Cheng Y, Jiang Y, Liu S, Zhang M, Liu J, et al. Ten hub genes associated with progression and prognosis of pancreatic carcinoma identified by co-expression analysis. Int J Biol Sci. 2018;14(2):124–36.
Li T, Wernersson R, Hansen RB, Horn H, Mercer J, Slodkowicz G, et al. A scored human protein–protein interaction network to catalyze genomic interpretation. Nat Methods. 2017;14(1):61–4.
Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171(6):1437.
Wagner AH, Coffman AC, Ainscough BJ, Spies NC, Skidmore ZL, Campbell KM, et al. DGIdb 2.0: mining clinically relevant drug–gene interactions. Nucleic Acids Res. 2016;44(D1):D1036–44.
Wang Y, Zeng T. Response to: practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2013;14(1):391.
Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–5.
Chen Y, Liang Y, Chen Y, Ouyang S, Liu K, Yin W. Identification of prognostic metabolism-related genes in clear cell renal cell carcinoma. J Oncol. 2021;2021:2042114.
Li J, Yang Z-L, Ren X, Zou Q, Yuan Y, Liang L, et al. ACE2 and FZD1 are prognosis markers in squamous cell/adenosquamous carcinoma and adenocarcinoma of gallbladder. J Mol Histol. 2014;45(1):47–57.
Li Y, Liu Z, Zhang Y. Expression and prognostic impact of FZDs in pancreatic adenocarcinoma. BMC Gastroenterol. 2021;21(1):1–14.
Peng Q, Wang L, Zhao D, Lv Y, Wang H, Chen G, et al. Overexpression of FZD1 is associated with a good prognosis and resistance of sunitinib in clear cell renal cell carcinoma. J Cancer. 2019;10(5):1237–51.
Su W, Ye H, Wu F, Mo Y, Li S, Mai Z, et al. Low expression of FZD1 correlates with poor prognosis in non-small cell lung cancer. Int J Clin Exp Pathol. 2016;9(8):8335–42.
Yang L, Yang Z, Li D, Liu Z, Zou Q, Yuan Y, et al. Overexpression of FZD1 and CAIX are associated with invasion, metastasis, and poor-prognosis of the pancreatic ductal adenocarcinoma. Pathol Oncol Res. 2018;24(4):899–906.
Chang VHS, Chu P-Y, Peng S-L, Mao T-L, Shan Y-S, Hsu C-F, et al. Kruppel-like factor 10 expression as a prognostic indicator for pancreatic adenocarcinoma. Am J Pathol. 2012;181(2):423–30.
Lin Y-M, Yeh K-T, Yeh C-M, Soon M-S, Hsu L-S. KLF10 functions as an independent prognosis factor for gastric cancer. Medicina Lithuania. 2022;58(6):711.
Yeh C-M, Lee Y-J, Ko P-Y, Lin Y-M, Sung W-W. High expression of KLF10 is associated with favorable survival in patients with oral squamous cell carcinoma. Medicina Lithuania. 2021;57(1):17.
Zhao Z, Zhan Y, Jing L, Zhai H. KLF10 upregulates ACSM3 via the PI3K/Akt signaling pathway to inhibit the malignant progression of melanoma. Oncol Lett. 2022;23(6):1–11.
Ferraro B, Bepler G, Sharma S, Cantor A, Haura EB. EGR1 predicts PTEN and survival in patients with non-small-cell lung cancer. J Clin Oncol. 2005;23(9):1921–6.
Cloughesy TF, Cavenee WK, Mischel PS. Glioblastoma: from molecular pathology to targeted treatment. In: Abbas AK, Galli SJ, Howley PM, editors. Annual review of pathology: mechanisms of disease, Vol 9. Annual review of pathology-mechanisms of disease. 92014. p. 1–25.
Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–96.
Zanca C, Villa GR, Benitez JA, Thorne AH, Koga T, D’Antonio M, et al. Glioblastoma cellular cross-talk converges on NF-kappa B to attenuate EGFR inhibitor sensitivity. Genes Dev. 2017;31(12):1212–27.
Zhao L, Wang W, Xu L, Yi T, Zhao X, Wei Y, et al. Integrative network biology analysis identifies miR-508-3p as the determinant for the mesenchymal identity and a strong prognostic biomarker of ovarian cancer (vol 38, pg 2305, 2019). Oncogene. 2019;38(47):7279–80.
Lu D-H, Yang J, Gao L-K, Min J, Tang J-M, Hu M, et al. Lysine demethylase 2A promotes the progression of ovarian cancer by regulating the PI3K pathway and reversing epithelial-mesenchymal transition. Oncol Rep. 2019;41(2):917–27.
Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–9.
Miao L, Zhang Y, Huang L. mRNA vaccine for cancer immunotherapy. Mol Cancer. 2021;20(1):1–23.
Grabacka M, Wieczorek J, Michalczyk-Wetula D, Malinowski M, Wolan N, Wojcik K, et al. Peroxisome proliferator-activated receptor alpha (PPAR alpha) contributes to control of melanogenesis in B16 F10 melanoma cells. Arch Dermatol Res. 2017;309(3):141–57.
Suyama K, Naito Y, Yoh K, Niho S, Goto K, Ohmatsu H, et al. Development of cushing’s syndrome during effective chemotherapy for small cell lung cancer. Intern Med. 2011;50(4):335–8.
Russell G, Lightman S. The human stress response. Nat Rev Endocrinol. 2019;15(9):525–34.
Latour M, Her N-G, Kesari S, Nurmemmedov E. WNT signaling as a therapeutic target for glioblastoma. Int J Mol Sci. 2021;22(16):8428.
Jung Y-S, Park J-I. Wnt signaling in cancer: therapeutic targeting of Wnt signaling beyond beta-catenin and the destruction complex. Exp Mol Med. 2020;52(2):183–91.
Kalani MYS, Cheshier SH, Cord BJ, Bababeygy SR, Vogel H, Weissman IL, et al. Wnt-mediated self-renewal of neural stem/progenitor cells. Proc Natl Acad Sci. 2008;105(44):16970–5.
Chen Q, Cai J, Wang Q, Wang Y, Liu M, Yang J, et al. Long noncoding RNA NEAT1, regulated by the EGFR pathway, contributes to glioblastoma progression through the WNT/β-catenin pathway by scaffolding EZH2. Clin Cancer Res. 2018;24(3):684–95.
Suwala AK, Hanaford A, Kahlert UD, Maciaczyk J. Clipping the wings of glioblastoma: modulation of WNT as a novel therapeutic strategy. J Neuropathol Exp Neurol. 2016;75(5):388–96.
Sareddy GR, Kesanakurti D, Kirti PB, Babu PP. Nonsteroidal anti-inflammatory drugs diclofenac and celecoxib attenuates Wnt/beta-catenin/Tcf signaling pathway in human glioblastoma cells. Neurochem Res. 2013;38(11):2313–22.
Yin YQ, Cai JJ, Meng FD, Sui CG, Jiang YH. MiR-144 suppresses proliferation, invasion, and migration of breast cancer cells through inhibiting CEP55. Cancer Biol Ther. 2018;19(4):306–15.
Liborio-Kimura TN, Jung HM, Chan EKL. miR-494 represses HOXA10 expression and inhibits cell proliferation in oral cancer. Oral Oncol. 2015;51(2):151–7.
Bi W, Huang JY, Nie CL, Liu B, He GQ, Han JH, et al. CircRNA circRNA_102171 promotes papillary thyroid cancer progression through modulating CTNNBIP1-dependent activation of -catenin pathway. J Exp Clin Cancer Res. 2018;37:1–9.
Barbagallo D, Condorelli A, Ragusa M, Salito L, Sammito M, Banelli B, et al. Dysregulated miR-671-5p/CDR1-AS/CDR1/VSNL1 axis is involved in glioblastoma multiforme. Oncotarget. 2016;7(4):4746–59.
Kurscheid S, Bady P, Sciuscio D, Samarzija I, Shay T, Vassallo I, et al. Chromosome 7 gain and DNA hypermethylation at the HOXA10 locus are associated with expression of a stem cell related HOX-signature in glioblastoma. Genome Biol. 2015;16:1–15.
Li F, Jin D, Guan L, Zhang C-C, Wu T, Wang Y-J, et al. CEP55 promoted the migration, invasion and neuroshpere formation of the glioma cell line U251. Neurosci Lett. 2019;705:80–6.
Amaral RF, Geraldo LHM, Einicker-Lamas M, Spohr TCLDSE, Mendes F, Lima FRS. Microglial lysophosphatidic acid promotes glioblastoma proliferation and migration via LPA(1)receptor. J Neurochem. 2021;156(4):499–512.
Wu F, Jiao J, Liu F, Yang Y, Zhang S, Fang Z, et al. Hypermethylation of Frizzled1 is associated with Wnt/beta-catenin signaling inactivation in mesenchymal stem cells of patients with steroid-associated osteonecrosis. Exp Mol Med. 2019;51:1–9.
Han SY, Pang MF, Nelson CM. Substratum stiffness tunes proliferation downstream of Wnt3a in part by regulating integrin-linked kinase and frizzled-1. J Cell Sci. 2018;131(8):210476.
Planutis K, Planutiene M, Nguyen AV, Moyer MP, Holcombe RF. Invasive colon cancer, but not non-invasive adenomas induce a gradient effect of Wnt pathway receptor frizzled 1 (Fz1) expression in the tumor microenvironment. J Transl Med. 2013;11:1–7.
Flahaut M, Meier R, Coulon A, Nardou KA, Niggli FK, Martinet D, et al. The Wnt receptor FZD1 mediates chemoresistance in neuroblastoma through activation of the Wnt/beta-catenin pathway. Oncogene. 2009;28(23):2245–56.
Zhang H, Jing X, Wu X, Hu J, Zhang X, Wang X, et al. Suppression of multidrug resistance by rosiglitazone treatment in human ovarian cancer cells through downregulation of FZD1 and MDR1 genes. Anticancer Drugs. 2015;26(7):706–15.
Zhang H, Zhang X, Wu X, Li W, Su P, Cheng H, et al. Interference of Frizzled 1 (FZD1) reverses multidrug resistance in breast cancer cells through the Wnt/beta-catenin pathway. Cancer Lett. 2012;323(1):106–13.
Memon A, Lee WK. KLF10 as a tumor suppressor gene and its TGF-beta signaling. Cancers. 2018;10(6):161.
Hsu Y-C, Ho C, Shih Y-H, Ni W-C, Li Y-C, Chang H-C, et al. Knockout of KLF10 ameliorated diabetic renal fibrosis via downregulation of DKK-1. Molecules. 2022;27(9):2644.
You M, Zhang L, Zhang X, Fu Y, Dong X. MicroRNA-197-3p inhibits the osteogenic differentiation in osteoporosis by down-regulating KLF 10. Clin Interv Aging. 2021;16:107–17.
Papadakis KA, Krempski J, Svingen P, Xiong Y, Sarmento OF, Lomberk GA, et al. Kruppel-like factor KLF10 deficiency predisposes to colitis through colonic macrophage dysregulation. Am J Physiol Gastrointest Liver Physiol. 2015;309(11):G900–9.
Zhou MM, Chen JQ, Zhang H, Liu HL, Yao H, Wang XM, et al. KLF10 inhibits cell growth by regulating PTTG1 in multiple myeloma under the regulation of microRNA-106b-5p. Int J Biol Sci. 2020;16(12):2063–71.
Marrero-Rodriguez D, Taniguchi-Ponciano K, Subramaniam M, Hawse JR, Pitel KS, Arreola-DelaCruz H, et al. Kruppel-like factor 10 participates in cervical cancer immunoediting through transcriptional regulation of pregnancy-specific beta-1 glycoproteins. Sci Rep. 2018;8:9445.
Diamond JR, Becerra C, Richards D, Mita A, Osborne C, O’Shaughnessy J, et al. Phase Ib clinical trial of the anti-frizzled antibody vantictumab (OMP-18R5) plus paclitaxel in patients with locally advanced or metastatic HER2-negative breast cancer. Breast Cancer Res Treat. 2020;184(1):53–62.
Davis SL, Cardin DB, Shahda S, Lenz HJ, Dotan E, O’Neil BH, et al. A phase 1b dose escalation study of Wnt pathway inhibitor vantictumab in combination with nab-paclitaxel and gemcitabine in patients with previously untreated metastatic pancreatic cancer. Invest New Drugs. 2020;38(3):821–30.
Wieland A, Trageser D, Gogolok S, Reinartz R, Höfer H, Keller M, et al. Anticancer effects of niclosamide in human glioblastoma. Clin Cancer Res. 2013;19(15):4124–36.
Cheng B, Morales LD, Zhang Y, Mito S, Tsin A. Niclosamide induces protein ubiquitination and inhibits multiple pro-survival signaling pathways in the human glioblastoma U-87 MG cell line. PLoS ONE. 2017;12(9): e0184324.
Oh HC, Shim JK, Park J, Lee JH, Choi RJ, Kim NH, et al. Combined effects of niclosamide and temozolomide against human glioblastoma tumorspheres. J Cancer Res Clin Oncol. 2020;146(11):2817–28.
Kumar SU, Gopinath P. Controlled delivery of bPEI-niclosamide complexes by PEO nanofibers and evaluation of its anti-neoplastic potentials. Colloids Surf B Biointerfaces. 2015;131:170–81.
Kraljic K, Jelic D, Ziher D, Cvrtila A, Dragojevic S, Sinkovic V, et al. Benzoxaboroles-novel autotaxin inhibitors. Molecules. 2019;24(19):3419.
Acknowledgements
We appreciate the online resources of public databases such as the GO and KEGG etc.
Funding
This study was funded by the Xiamen Municipal Bureau of Science and Technology (3502Z20209005), Natural Science Foundation of Fujian Province (2020J02063), and the National Natural Science Foundation of China (82072777), the Fujian Provincial Clinical Research Center for Brain Diseases (2021FJSLCYX01). The funders had no role in study design, data collection, analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
FJ and LZ wrote the main manuscript text, analyzed the data, and contributed reagents, materials, and analysis tools. ZJ performed the experiments and prepared figures or tables. GT and ZW reviewed drafts of the paper, and approved the final draft. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All methods were carried out under relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
There are no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
. The supplementary tables of this study.
Additional file 2
. The supplementary figures of this study.
Additional file 3
. Some of the scripts used in this study.
Additional file 4
. The statistical characteristics of DEmRNAs identified in this study.
Additional file 5
. The P-values and log2 FC of DEmiRNAs identified in this study.
Additional file 6
. The specific information of DEcircRNAs identified in this study.
Additional file 7
. The statistical characteristics of DEcircRNAs in this study.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Jia, F., Zhang, L., Jiang, Z. et al. FZD1/KLF10-hsa-miR-4762-5p/miR-224-3p-circular RNAs axis as prognostic biomarkers and therapeutic targets for glioblastoma: a comprehensive report. BMC Med Genomics 16, 21 (2023). https://doi.org/10.1186/s12920-023-01450-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12920-023-01450-w