Abstract
Background
Antimicrobial resistance (AMR) is widely acknowledged as a global problem, yet in many parts of the world its magnitude is still not well understood. This review, using a public health focused approach, aimed to understand and describe the current status of AMR in Africa in relation to common causes of infections and drugs recommended in WHO treatment guidelines.
Methods
PubMed, EMBASE and other relevant databases were searched for recent articles (2013–2016) in accordance with the PRISMA guidelines. Article retrieval and screening were done using a structured search string and strict inclusion/exclusion criteria. Median and interquartile ranges of percent resistance were calculated for each antibiotic-bacterium combination.
Results
AMR data was not available for 42.6% of the countries in the African continent. A total of 144 articles were included in the final analysis. 13 Gram negative and 5 Gram positive bacteria were tested against 37 different antibiotics. Penicillin resistance in Streptococcus pneumoniae was reported in 14/144studies (median resistance (MR): 26.7%). Further 18/53 (34.0%) of Haemophilus influenza isolates were resistant to amoxicillin. MR of Escherichia coli to amoxicillin, trimethoprim and gentamicin was 88.1%, 80.7% and 29.8% respectively. Ciprofloxacin resistance in Salmonella Typhi was rare. No documented ceftriaxone resistance in Neisseria gonorrhoeae was reported, while the MR for quinolone was 37.5%. Carbapenem resistance was common in Acinetobacter spp. and Pseudomonas aeruginosa but uncommon in Enterobacteriaceae.
Conclusion
Our review highlights three important findings. First, recent AMR data is not available for more than 40% of the countries. Second, the level of resistance to commonly prescribed antibiotics was significant. Third, the quality of microbiological data is of serious concern. Our findings underline that to conserve our current arsenal of antibiotics it is imperative to address the gaps in AMR diagnostic standardization and reporting and use available information to optimize treatment guidelines.
Similar content being viewed by others
Background
Internationally, there is a growing concern over antimicrobial resistance (AMR) which is currently estimated to account for more than 700,000 deaths per year worldwide [1]. If no appropriate measures are taken to halt its progress, AMR will cost approximately 10 million lives and about US$100 trillion per year by 2050 [1]. In contrast to some other health issues, AMR is a problem that concerns every country irrespective of its level of income and development as resistant pathogens do not respect borders [1, 2].
Despite the threat presented by AMR, the 2014 World Health Organization (WHO) and the recent O’Neill report describe significant gaps in surveillance, standard methodologies and data sharing [1, 2]. The 2014 WHO report identified Africa and South East Asia as the regions without established AMR surveillance systems [2]. This lack of quality data is problematic often leading to treatment guidelines that are not adequate for the local situation. The gap in public health capacity is also an issue given the changing resistance mechanisms and the emergence of multidrug-resistant bacteria that can only be detected through systematic screening in quality assured microbiology laboratories [3, 4].
One factor contributing to AMR is misuse of antibiotics. Improved malaria diagnostics and the recognition that malaria transmission is decreasing globally has highlighted the lack of tests for other infections and many patients who test negative for malaria are treated with antibiotics indiscriminately [5,6,7]. Clinical treatment algorithms like the Integrated Management of Neonatal and Childhood Illnesses (IMNCI) and Integrated Management of Adolescent and Adulthood Illnesses (IMAI) guidelines implemented by the WHO have tried to optimize antibiotic prescription in resource-limited settings, however overuse of antibiotics is still happening [8,9,10,11]. Following these guidelines amoxicillin or sulfamethoxazole/trimethoprim are the first line drugs for urinary tract infections (UTI) or acute respiratory tract infections. A combination of ampicillin and gentamicin or ceftriaxone are the drugs of choice for treating blood stream infections (BSI) and sulfamethoxazole/trimethoprim or ciprofloxacin are recommended for the treatment of dysentery [8, 9, 12].
A number of recent reviews summarized AMR data in Africa, most recently Leopold et al. (2014) which focused on Sub-Saharan Africa. The authors found a high level of resistance to the commonly used antibiotics in the sub-Saharan African region. For example, 90% of Gram negatives were resistant to chloramphenicol, a commonly used antibiotic. In contrast, resistance to third-generation cephalosporins (like ceftriaxone) was less common, recommending this group for use [13].
To design suitable local and global interventions, it is important to understand the current status of AMR and identify knowledge gaps. The purpose of this review is to summarize the available information about the occurrence of AMR on the entire African continent and describe laboratory methods currently in use, to identify knowledge gaps and highlight diagnostic needs.
Methods
Search strategy
PubMed, EMBASE, Science Daily, the Cochrane Database for Systematic Reviews, African Journals Online Library and Free-text Web Searches using Google Scholar were searched for articles published in English from January 1, 2013 through January 31, 2016. Literature before January 2013 was covered in previous reviews [13, 14]. Reference lists of relevant articles were checked for additional titles for inclusion in the review. Key words used for the search were “Antimicrobial Resistance”, “Antimicrobial Susceptibility”, “Surveillance”, “Diagnostic”, “Africa” and specific names of all African countries. The detailed search strategy, as well as details of the article quality assessment can be found in Additional file 1.
Selection criteria
Articles reporting AMR prevalence, availability of AMR surveillance systems or diagnostic needs of antibiotic resistance in the whole African region were included. Based on the abstract, articles of all types with any data on etiology and antibiotic susceptibility pattern were included for further screening. Studies were included or excluded following predefined criteria.
Inclusion:
-
Reports on AMR in humans from the African region
-
Abstracts and full text available in English
-
Drug sensitivity testing done in a laboratory setting with defined cutoffs for drug susceptibility testing
-
The denominator as total isolates clearly described for population based studies
-
Case reports and case series
Exclusion:
-
Reports published before 2013
-
Studies only focused on malaria, HIV or tuberculosis without AMR information
-
Studies without information on total studied isolates
Selection procedure
Titles and abstracts of all the articles retrieved through the search were screened. In the event of uncertainty as to whether articles met the criteria for study inclusion they were discussed with two co- authors. Articles selected for full text review were obtained using PubMed, WHO GIFT access, HINARI, institutional websites or by contacting the authors directly. Names of authors from articles in the search results were not blinded for abstract or full-text review.
Data extraction
Data extraction was done using a predesigned and pretested database, developed for the purposes of this review using Microsoft Excel 2013. Information extracted included article information (PMID, first author, year of publication, duration of data collection and country), study design (sample size, age group, hospital acquired or community acquired, number of specimens collected, and clinical syndrome), pathogen identification and antimicrobial susceptibility testing methodology, laboratory accreditation information and antibacterial resistance data.
Article quality assessment
The quality of each article was assessed using a tool modified for the purposes of this study from criteria published by Omulo et al. and the Cochrane guidelines for assessing bias in observational studies. Since a limited number of articles was available, results of the quality assessment were not used for inclusion/exclusion. The quality criteria included 26 items to assess the design, details of sample collection, processing and storage, reporting on AMR methodologies and quality assurance strategies.
Data analysis
We calculated prevalence, median resistance (MR) and inter quartile range (IQR) of resistance for each bacterium-antibiotic combination to calculate a standardized measure from the collected data. Pediatric age was considered less than 18 years and neonatal age less than 28 days. Meta-analysis was not conducted because of the large variability in AMR methodology, geography and the small number of articles available per country. Since the number of studies from hospital/in-patient settings was small, they were combined and median percentages with interquartile ranges were generated. Statistical analyses and visualization were performed using Microsoft Excel 2013, STATA v14 (STATA, College Station, TX, USA) and R-software 3.3.1.
Results
Data and study characteristics
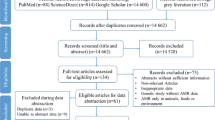
In total, 1704 articles were identified. Of those, 144 studies met the inclusion criteria and were included in the final analysis (Fig. 1). Samples from a total of 149,733 patients were analyzed in the selected studies. The majority of the studies were from East Africa (59/144, 40.9%) while the smallest number of studies were from the South African region (6/144, 4.2%) (Fig. 2). No suitable report was identified from 23/54, 42.6% countries. While the articles were published between 2013 and 2016, the reported data were collected from 1995 to 2015 with the majority from before 2013 (98/144, 68.1%). Most of the studies (92/144, 63.9%) were cross sectional studies or case series (Table 1). Similar numbers of studies were published with susceptibility data for isolates from blood culture (25/144, 17.4%), urine culture (25/144, 17.4%), wound discharge/pus isolates (22/144, 15.3%); and multiple sample types (21/144, 14.6%). More than 80% of the studies fulfilled more than half of the quality parameters (121/144, 84.0%) used to score the articles (Additional file 1). Among the different studies, four different methods for susceptibility testing and five different interpretation guidelines were used (Table 1).
PRISMA Diagram of the article selection procedure for articles published between January 2013 and January 2016.The review has been registered to the PROSPERO database of systematic reviews on March 1, 2016 (http://www.crd.york.ac.uk/PROSPERO/myprospero.php) with ID CRD42016035923
Geographical distribution and number of selected studies between January 2013 and January 2016 in the different African countries. Countries were grouped based on the United Nations Statistics Division classification into Eastern Africa, Southern Africa, Central Africa, Northern Africa and Western Africa
Microbial resistance patterns
The most commonly reported bacterium was Escherichia coli (87/144, 60.4%) with the most frequent susceptibility data for gentamicin (77/144, 53.5%), ciprofloxacin (71/144, 49.3%) and sulfamethoxazole/trimethoprim (68/144, 47.2%) (Table 2). In total, 13 Gram negative opportunistic pathogens were tested against 37 antibiotics (Table 3, Additional file 2). Overall resistance to commonly used drugs [8, 9, 12], like amoxicillin (MR 72.9%, IQR9.1%–87.3%) and trimethoprim/sulfamethoxazole (MR 75.0%, IQR 49.5%–92.3%) was high. Low to moderate resistance was found to gentamicin (MR 22.1%, IQR 2.0%–45.0%), ciprofloxacin (MR 16.7%, IQR 0%–38.5%) and ceftriaxone (MR 17.2%, IQR 0%-45.0%). Gentamicin resistance in Klebsiella spp., which is naturally resistant to ampicillin, was reported in 1031/2715, 38.0% of tested isolates. Resistance to either ceftriaxone or cefotaxime, which is suggestive of extended–spectrum beta lactamase (ESBL) production was reported in 593/2963, 20.0% and 1051/5395, 19.5% of E. coli as well as 545/1594, 34.2% and 560/1199, 46.7% of the K. pneumoniae isolates, respectively. Imipenem, a drug rarely available in rural Africa, was investigated in 21/144 (14.6%) of the studies and showed the lowest overall MR (MR 3.0%, IQR 0%- 26.6%) (Table 3). Of the Acinetobacter isolates reported in three studies, 32.3% (10/31) were resistant to meropenem. Ciprofloxacin resistance in S. Typhi was reported rarely (MR 0%, IQR 0%–11.7%) while ampicillin resistance in Haemophilus influenza was high (MR 100%, IQR76.6%–100%). There was no documented ceftriaxone resistance in N. gonorrhoeae but ciprofloxacin resistance was reported in 37.5% (IQR 30.7%–83.9%) of the isolates.
In the Gram positive group, Coagulase Negative Staphylococcus spp. (CoNS), S. aureus, Streptococcus pneumoniae and Group A streptococcus were the most commonly investigated bacteria (Table 4). Methicillin resistant S. aureus (MRSA) was reported in 7/79 (8.9%) of the studies, however as cefoxitin is typically used to screen for MRSA, the MRSA rate is likely underestimated [15]. Erythromycin resistance in S. aureus was found in 33.9% (IQR 13%–46.4%). Methods used to assess penicillin susceptibility in S. pneumoniae varied: median oxacillin resistance was reported in 40.7% (IQR 0%–55.7%) compared to 26.7% (IQR 8.4%–33.6%) for penicillin and 22.5% (IQR 20%–32.5%) for amoxicillin.Vancomycin showed the lowest resistance pattern for all the tested Gram positive bacteria (Table 4).
In the reviewed literature, E. coli was commonly isolated from patients with BSI (17/87, 19.5%), UTI (17/87, 19.5%) and wound infection (16/87, 18.4%). Citrobacter spp. were reported most commonly among patients with UTIs (7/19; 36.8%), followed by BSI (5/19, 26.3%). Two (2/19, 10.6%) [16, 17] of the studies which reported on Citrobacter spp. were from HAI; five (5/19, 26.3%) from community acquired infections of which three (3/5) came from pregnant women at antenatal care follow up [18,19,20] and two (2/5) had unknown risk factors [21, 22]. S. Typhi was most frequently reported from patients with BSI (13/28, 46.4%) followed by patients with diarrhea (6/28, 21.4%). It was also more commonly reported in children below 18 years (12/28, 48.9%) than in adults (2/28, 7.1%) (Table 2).
In the current review, the susceptibility results were taken at face value, however there were inconsistencies casting the credibility of several results in doubt. For example a study reporting more than 80% of Proteus isolates (n = 6) as resistant to imipenem also reported that 80% of them were susceptible to ceftriaxone and 50% were susceptible to vancomycin, a combination that is highly unusual [23]. There were other unverified reports of highly unusual resistance patterns from some centers, such as penicillin resistant S. pyogenes [24, 25] and vancomycin-resistant S. aureus [26, 27].
Regional antibiotic resistance patterns
Generally, a lower level of resistance of S. aureus, Klebsiella spp., E. coli and S. pneumoniae to carbapenems and fluoroquinolones was observed in all the regions as compared to the other antibiotic-bacterium combinations. However, Klebsiella spp. resistance to ciprofloxacin in West Africa was observed to be higher than in other regions. Resistance to the trimethoprim (MR: 33.9%–100%), ampicillin (MR: 7.9%–100%) and penicillin (MR: 0%–75%) was generally high in all regions (Fig. 3).
Resistance of selected pathogens to commonly prescribed antibiotics in different regions of Africa. The boxplots in the figure represent the median and interquartile range of resistance reported if at least three studies reported on the combination. Resistance to amoxicillin-clavulanic acid (AMX-CLA), ampicillin, amoxicillin, penicillin, oxacillin, trimethoprim-sulfamethoxazole (TMP-SXT), gentamicin, ceftriaxone, cefoxitin, ciprofloxacin, erythromycin, tetracycline, vancomycin and imipenem were plotted. Antibiotics with no data points in the specific regions are omitted from the plots.Resistance patterns reported using broth dilution minimum inhibitory concentration (MIC), E-test® or VITEK® were included if prevalence could be calculated and were combined with resistance data reported using disk diffusion as this was the main method used. Intermediate susceptible strains were categorized as resistant to simplify the analysis. [13]MR estimates were not calculated if only one or two studies reported on the specific bacterium-antibiotic combination. a: Resistance of Klebsiella spp. to commonly prescribed antibiotics in different regions of Africa. b: Resistance of S. pneumoniae to commonly prescribed antibiotics in different regions of Africa. c: Resistance of S. aureus to commonly prescribed antibiotics in different regions of Africa. d: Resistance of E. coli to commonly prescribed antibiotics in different regions of Africa
Discussion
The rise and spread of AMR threatens the effective control and treatment of various bacterial diseases worldwide [1, 2]. The achievements gained in reducing mortality and morbidity through early use of antibiotics based on empiric guidelines are in serious jeopardy if appropriate actions are not taken to control AMR [28, 29]. Availability of routine and research data on pathogen susceptibilities is an important step towards designing targeted strategies to tackle the global AMR crisis. The current review describes recently (2013–2016) published data on antibiotic drug susceptibility from Africa.
The lack of consistency in the measurement and reporting of susceptibility data makes it difficult to compare findings among different countries and laboratories, sometimes even within one country [30, 31]. To address this issue, high income countries have implemented harmonization efforts. For example, laboratories in Europe are encouraged to use the European Committee on Antimicrobial Susceptibility Testing (EUCAST) standard over the Clinical and Laboratory Standards Institute (CSLI) guidelines [32]. Furthermore, in an effort to enable coherent data synthesis and reporting in January 2016 the British Society for Antimicrobial Chemotherapy (BSAC) actively promoted the EUCAST methods in favour of the current BSAC methodology [33]. Given the findings of our review, similar harmonization efforts are urgently needed in Africa. Standardizing AMR methods and interpretation guidelines could allow for better comparability of results and improved resistance tracking. Furthermore, improved access to reference laboratories and EQA schemes are needed augmenting the current WHO initiative to scale up the global antimicrobial surveillance system (GLASS) based on country specific priority pathogens [34]. Currently, in the absence of a uniform laboratory methodology the GLASS goals will be very difficult to meet.
Comparing our findings with previous reviews in the region like Leopold et al. (2014), overall, we identify a similarly high report of resistance to commonly used antibiotics [13]. The same review reported a high level of resistance of Enterobacteriaceae to ampicillin and co-trimoxazole which is in agreement with our findings. Similarly, resistance to co-trimoxazole and tetracycline by S. pneumoniae was reported to be high. However, discrepancies were observed in various antibiotic/bacterium combinations. For instance, our finding of resistance to chloramphenicol in Salmonella Typhi isolates is lower than that previously reported [13]. Resistance to oxacillin by S. aureus was also much higher in the current review than the review by Leopold et al. [13]. The observed differences between data published in 2014 and the current work could indicate a rising pattern in AMR in certain pathogens. However it could also be because of the differences in AMR testing methodologies underlining the need for harmonization of laboratory methods in the region.
A notable finding of this review was the high resistance rate of common causes of UTI to common first line regimes like amoxicillin and sulfamethoxazole/trimethoprim. In the presence of a failing treatment, patients with UTIs are at increased risk of developing renal damage and future risks of renal insufficiency or hypertension [35]. Similarly, given the resistance profiles in the current review, neonatal sepsis or BSI caused by E. coli, K. pneumoniae and S. aureus are not being effectively treated by first line drugs like ampicillin, aminoglycosides and cephalosporins, which will result in increased mortality in patients with life-threatening infections. The high levels of resistance to amoxicillin and penicillin in S. pneumoniae and H. influenzae are also concerning given that pneumonia is a leading cause of death in children [36, 37]. Reported MRSA rates were variable and doubts remain about reliability of identification at all sites which are confirmed by the findings of one Kenyan study which found that MRSA rates dropped dramatically after switching to an automated identification method [38].
Compared to reports from Asia, quinolone resistance in S. Typhi was rare and, reassuringly, there were no reports of ceftriaxone resistance in N. gonorrhoeae [39]. Less commonly prescribed antibiotics like imipenem and vancomycin also showed low level resistance and they should be preserved as alternative drugs in severe infections. Most of the imipenem resistance was described in isolates of P. aeruginosa which has been reported from other centers [33]. Oxacillin resistance should predict penicillin resistance in S. pneumoniae reliably [40, 41], however in the current review resistance to oxacillin was much higher than to penicillin, possibly because of the differences in the number of isolates tested for both antibiotics and the use of different cut-offs for meningitis and non-meningitis strains .
The results also yielded data on the susceptibility of less commonly described bacteria like Acinetobacter spp. and Citrobacter spp. Acinetobacter spp. are especially important given their importance in clinical infections and the reported rising trend of resistance, further they have been included in the priority pathogens for global surveillance based on the GLASS initiative [42]. Citrobacter spp. were reported from hospital and community settings, including in pregnant mothers and can cause UTI which puts pregnant women at risk of preterm labor. Our results would suggest that current frontline treatments are ineffective against most common uro-pathogens.
The limitations of the current review include the exclusion of non-English language reports, as articles from French speaking African countries might have been missed, biasing this review. The representativeness of the data is hard to assess as it is possible that the absence of resistance is not routinely reported and focus is given to reports of resistance. There were very few reports from South Africa, which has a better functioning health system than neighboring countries and better national AMR surveillance. These data were not accessible by our search and therefore larger AMR trends might have been missed. A further limitation is combining AMR results from different patient groups across different countries to compare the data. This approach might have leveled out peaks of resistance in different settings. However, given the observed trends, we believe that the resolution of the obtained data was sufficient to show general developments. Moreover, since case reports were not excluded with intention of capturing as much data as possible in the current review, the findings from the three case (3/144, 2.1% of the studies) might bias the resistance testing results as case reports tend to report on specific multi-drug resistant pathogens. Finally, resistance data obtained with different laboratory methodologies were combined for the purposes of this review. However, as the majority of studies used the disk diffusion method and CLSI guidelines, the impact of the variation in AMR methodology on the validity of the final results is thought to be minimal.
Conclusion
In summary, our review highlights three important findings: first, more than a third of the countries on the continent did not have recent AMR data published in the public domain and only a few of those were surveillance data. Second, a high level of drug resistance exists to commonly prescribed antibiotics in the African continent. Third, the standardization and quality of the microbiological identification and susceptibility testing methods needs to be improved to allow national and international organizations to monitor the extent of the AMR problem. All of the identified areas of concern need urgent attention by the global health community in order to halt the public health threat associated with spreading AMR.
References
O’Neill J: Tackling Drug-Resistant Infections Globally: Final Report and Recommendations May 2016(http://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf).
WHO: Antimicrobial Resistance. In: Global Report on surveillance. Edited by WHO. Geneva, Switzerland; 2014.
Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–8.
Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, Malhotra-Kumar S. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia Coli, Belgium, June 2016. Euro Surveill. 2016;21(27)
WHO: World Malaria Report. In. Edited by WHO, vol. http://apps.who.int/iris/bitstream/10665/200018/1/9789241565158_eng.pdf?ua=1. Switzerland, Geneva; 2015.
Harbarth S, Samore MH. Antimicrobial resistance determinants and future control. Emerg Infect Dis. 2005;11(6):794–801.
Do NT, Ta NT, Tran NT, Than HM, Vu BT, Hoang LB, van Doorn HR, Vu DT, Cals JW, Chandna A, et al. Point-of-care C-reactive protein testing to reduce inappropriate use of antibiotics for non-severe acute respiratory infections in Vietnamese primary health care: a randomised controlled trial. Lancet Glob Health. 2016;
WHO: Integrated Managment of Neonatal and Childhood Illnesses (IMNCI). Switzerland, Geneva 2014, http://apps.who.int/iris/bitstream/10665/104772/16/9789241506823_Chartbook_eng.pdf?ua=1.
WHO: Integrated Managment of Adolescent and Adulthood Illnesses (IMAI). Switzerland, Geneva 2009, http://www.who.int/hiv/pub/imai/acute_care.pdf?ua=1.
Senn N, Rarau P, Salib M, Manong D, Siba P, Rogerson S, Mueller I, Genton B. Use of antibiotics within the IMCI guidelines in outpatient settings in Papua new Guinean children: an observational and effectiveness study. PLoS One. 2014;9(3):e90990.
Gwimile JJ, Shekalaghe SA, Kapanda GN, Kisanga ER. Antibiotic prescribing practice in management of cough and/or diarrhoea in Moshi municipality, Northern Tanzania: cross-sectional descriptive study. Pan Afr Med J. 2012;12:103.
WHO: Pocket book of hospital care for children: Second edition:Guidelines for the management of common childhood illnesses. Switzerland, Geneva 2013, http://www.who.int/maternal_child_adolescent/documents/child_hospital_care/en/.
Leopold SJ, van Leth F, Tarekegn H, Schultsz C. Antimicrobial drug resistance among clinically relevant bacterial isolates in sub-Saharan Africa: a systematic review. J Antimicrob Chemother. 2014;69(9):2337–53.
Le Doare K, Bielicki J, Heath PT, Sharland M. Systematic review of antibiotic resistance rates among gram-negative bacteria in children with sepsis in resource-limited countries. J Pediatric Infect Dis Soc. 2015;4(1):11–20.
Fernandes CJ, Fernandes LA, Collignon P. Cefoxitin resistance as a surrogate marker for the detection of methicillin-resistant Staphylococcus Aureus. J Antimicrob Chemother. 2005;55(4):506–10.
Mengesha RE, Kasa BG-S, Saravanan M, Berhe DF, Wasihun AG. Aerobic bacteria in post surgical wound infections and pattern of their antimicrobial susceptibility in Ayder teaching and referral hospital, Mekelle Ethiopia. BMC research notes. 2014;7:575.
Mohammed A, Adeshina GO, Ibrahim YK. Incidence and antibiotic susceptibility pattern of bacterial isolates from wound infections in a tertiary hospital in Nigeria. Trop J Pharm Res. 2013;12:617–21.
Tadesse E, Teshome M, Merid Y, Kibret B, Shimelis T. Asymptomatic urinary tract infection among pregnant women attending the antenatal clinic of Hawassa referral hospital Southern Ethiopia. BMC research notes. 2014;7:155.
Onoh RC, Umeora OUJ, Egwuatu VE, Ezeonu PO, Onoh TJP. Antibiotic sensitivity pattern of uropathogens from pregnant women with urinary tract infection in Abakaliki, Nigeria. Infect Drug Resist. 2013;6:225–33.
Oladeinde BH, Omoregie R, Oladeinde OB. Asymptomatic urinary tract infection among pregnant women receiving ante-natal care in a traditional birth home in Benin City Nigeria. Ethiop J Health Sci. 2015;25(1):3–8.
Wasihun AG, Wlekidan LN, Gebremariam SA, Dejene TA, Welderufael AL, Haile TD, Muthupandian S. Bacteriological profile and antimicrobial susceptibility patterns of blood culture isolates among febrile patients in Mekelle hospital Northern Ethiopia. SpringerPlus. 2015;4:314.
Abera B, Kibret M. Azithromycin, fluoroquinolone and chloramphenicol resistance of non-chlamydia conjunctival bacteria in rural community of Ethiopia. Indian J Ophthalmol. 2014;62:236–9.
Eida M, Nasser M, El-Maraghy N, Azab K. Pattern of hospital-acquired pneumonia in intensive care unit of Suez Canal university hospital Egyptian. Journal of Chest Diseases and Tuberculosis. 2015;64:625–31.
Muluye D, Wondimeneh Y, Moges F, Nega T, Ferede G. Types and drug susceptibility patterns of bacterial isolates from eye discharge samples at Gondar University Hospital, Northwest Ethiopia. BMC research notes. 2014;7:292.
Wasihun AG, Zemene Y. Bacterial profile and antimicrobial susceptibility patterns of otitis media in Ayder Teaching and Referral Hospital, Mekelle University, Northern Ethiopia. SpringerPlus. 2015;4:701.
Mengistu A, Gaeseb J, Uaaka G, Ndjavera C, Kambyambya K, Indongo L, Kalemeera F, Ntege C, Mabirizi D, Joshi MP, et al. Antimicrobial sensitivity patterns of cerebrospinal fluid (CSF) isolates in Namibia: implications for empirical antibiotic treatment of meningitis. Journal of Pharmaceutical Policy and Practice. 2013;6
Bayingana C. Antibiotic activity assessment of bacterial strains isolated from urine samples at Butare University Teaching Hospital (Buth) Laboratory. Int J Infect Dis. 2014;21:83.
Gera T, Shah D, Garner P, Richardson M, Sachdev HS. Integrated management of childhood illness (IMCI) strategy for children under five. Cochrane Database Syst Rev. 2016;6:CD010123.
Bhandari N, Mazumder S, Taneja S, Sommerfelt H, Strand TA. Effect of implementation of integrated Management of Neonatal and Childhood Illness (IMNCI) programme on neonatal and infant mortality: cluster randomised controlled trial. BMJ. 2012;344:e1634.
Perdigao-Neto LV, Oliveira MS, Rizek CF, Carrilho CM, Costa SF, Levin AS. Susceptibility of multiresistant gram-negative bacteria to fosfomycin and performance of different susceptibility testing methods. Antimicrob Agents Chemother. 2014;58(3):1763–7.
Lee M, Chung H-S. Different antimicrobial susceptibility testing methods to detect ertapenem resistance in enterobacteriaceae: VITEK2, MicroScan, Etest, disk diffusion, and broth microdilution. J Microbiol Methods. 2015;112:87–91.
ECDC. Surviellance report: annual report of the European antimicrobial resistance surveillance network (EARS-net). Sweden: Stockholm; 2014. http://ecdc.europa.eu/en/publications/Publications/antimicrobial-resistance-europe-2014.pdf
BSAC: Legacy BSAC Susceptibility Testing Method – no longer supported, for information only. http://www.bsacorguk/stewardship-surveillance/susceptibility/methodologylatestversion/.
WHO: Antimicrobial Resistance: Global Report on Surveillance. http://www.appswhoint/iris/bitstream/10665/188783/1/9789241549400_engpdf?ua=1 2015.
Park YS. Renal scar formation after urinary tract infection in children. Korean J Pediatr. 2012;55(10):367–70.
Woldu B, Bloomfield GS. Rheumatic heart disease in the twenty-first century. Curr Cardiol Rep. 2016;18(10):96.
Picot VS, Benet T, Messaoudi M, Telles JN, Chou M, Eap T, Wang J, Shen K, Pape JW, Rouzier V, et al. Multicenter case-control study protocol of pneumonia etiology in children: global approach to biological research, infectious diseases and epidemics in low-income countries (GABRIEL network). BMC Infect Dis. 2014;14:635.
Omuse G, Kabera B, Revathi G. Low prevalence of methicillin resistant Staphylococcus Aureus as determined by an automated identification system in two private hospitals in Nairobi Kenya: a cross sectional study. BMC Infect Dis. 2014;14:669.
Dalhoff A. Global fluoroquinolone resistance epidemiology and implictions for clinical use. Interdiscip Perspect Infect Dis. 2012;2012:976273.
Jette LP, Sinave C. Use of an oxacillin disk screening test for detection of penicillin- and ceftriaxone-resistant pneumococci. J Clin Microbiol. 1999;37(4):1178–81.
Gertrudis H, María LM, Liliana B, Sigri R, Luz C, Erik M, María EC, Rito Z, Eduardo C, Roger H, et al. Oxacillin disk diffusion testing for the prediction of penicillin resistance in Streptococcus Pneumoniae. Pan Am J Public Health. 2016;40(1):57–63.
Clark NM, Zhanel GG, Lynch JP 3rd. Emergence of antimicrobial resistance among Acinetobacter species: a global threat. Curr Opin Crit Care. 2016;22(5):491–9.
Aduda DSO, Macharia IM, Mugwe P, Oburra H, Farragher B, Brabin B, Mackenzie I. Bacteriology of chronic suppurative otitis media (CSOM) in children in Garissa district, Kenya: a point prevalence study. Int J Pediatr Otorhinolaryngol. 2013;77:1107–11.
Agmy G, Mohamed S, Gad Y, Farghally E, Mohammedin H, Rashed H. Bacterial profile, antibiotic sensitivity and resistance of lower respiratory tract infections in upper Egypt. Mediterranean Journal of Hematology and Infectious Diseases. 2013;5:1–7.
Ahmed EF, Gad GFM, Abdalla AM, Hasaneen AM, Abdelwahab SF. Prevalence of methicillin resistant Staphylococcus Aureus among Egyptian patients after surgical interventions. Surg Infect. 2014;15:404–11.
Ahmed MI, Alsammani MA, Babiker RA. Microbial profile in women with puerperal sepsis in Gadarif state, eastern Sudan. Annals of Tropical Medicine and Public Health. 2013;6:460–4.
Ahmed SF, Klena J, Husain T, Monestersky J, Naguib A, Wasfy MO. Genetic characterization of antimicrobial resistance of Shigella flexneri 1c isolates from patients in Egypt and Pakistan. Ann Clin Microbiol Antimicrob. 2013;12:9.
Akingbade OA, Olasunkanmi OI, Akinjinmi AA, Okerentugba PO, Onajobi BI, Okonko IO. Prevalence and antibiotic profile of Enterobacter species isolated from children with diarrhea in Abeokuta, Ogun state, Nigeria. Stem Cell. 2013;4:1–4.
Alabi AS, Frielinghaus L, Kaba H, Kosters K, Huson MAM, Kahl BC, Peters G, Grobusch MP, Issifou S, Kremsner PG, et al. Retrospective analysis of antimicrobial resistance and bacterial spectrum of infection in Gabon Central Africa. International Journal of Medical Microbiology. 2013;13:455.
Anagaw B, Gezachew M, Biadgelgene F, Anagaw B, Geleshe T, Taddese B, Getie B, Endris M, Mulu A, Unakal C. Antimicrobial susceptibility patterns of Streptococcus Pneumoniae over 6 years at Gondar University hospital, Northwest Ethiopia. Asian Pacific journal of tropical biomedicine. 2013;3:536–41.
Assefa A, Gelaw B, Shiferaw Y, Tigabu Z. Nasopharyngeal carriage and antimicrobial susceptibility pattern of streptococcus pneumoniae among pediatric outpatients at gondar university hospital, north west ethiopia. Pediatrics and Neonatology. 2013;54:315–21.
Babaiwa UF, Onyeagwara NC, Akerele JO. Bacterial tonsillar microbiota and antibiogram in recurrent tonsillitis. Biomedical Research (India). 2013;24:298–302.
Barogui YT, Klis S, Bankole HS, Sopoh GE, Mamo S, Baba-Moussa L, Manson WL, Johnson RC, van der Werf TS, Stienstra Y. Towards rational use of antibiotics for suspected secondary infections in Buruli ulcer patients. PLoS Negl Trop Dis. 2013;7:e2010.
Yeboah-Manu D, Kpeli GS, Ruf M-T, Asan-Ampah K, Quenin-Fosu K, Owusu-Mireku E, Paintsil A, Lamptey I, Anku B, Kwakye-Maclean C, et al. Secondary bacterial infections of buruli ulcer lesions before and after chemotherapy with streptomycin and rifampicin. PLoS Negl Trop Dis. 2013;7:e2191.
van der Meeren BT, Chhaganlal KD, Pfeiffer A, Gomez E, Ferro JJ, Hilbink M, Macome C, van der Vondervoort FJ, Steidel K, Wever PC. Extremely high prevalence of multi-resistance among uropathogens from hospitalised children in Beira, Mozambique. S Afr Med J. 2013;103:382–6.
Musiime V, Cook A, Bakeera-Kitaka S, Vhembo T, Lutakome J, Keishanyu R, Prendergast AJ, Lubwama S, Robertson V, Hughes P, et al. Bacteremia, causative agents and antimicrobial susceptibility among HIV-1-infected children on antiretroviral therapy in Uganda and Zimbabwe. Pediatr Infect Dis J. 2013;32:856–62.
Rasamiravaka T, Rasoanandrasana S, Zafindraibe NJJ, Rakoto Alson AO, Rasamindrakotroka A. Evaluation of methicillin-resistant Staphylococcus Aureus nasal carriage in Malagasy patients. J Infect Dev Ctries. 2013;7:318–22.
Kejela T, Bacha K. Prevalence and antibiotic susceptibility pattern of methicillin-resistant Staphylococcus Aureus (MRSA) among primary school children and prisoners in Jimma town Southwest Ethiopia. Ann Clin Microbiol Antimicrob. 2013;12:11.
Swierczewski BE, Odundo EA, Koech MC, Ndonye JN, Kirera RK, Odhiambo CP, Cheruiyot EK, Wu MT, Lee JE, Zhang C, et al. Surveillance for enteric pathogens in a case-control study of acute diarrhea in western Kenya. Trans R Soc Trop Med Hyg. 2013;107:83–90.
Sewunet T, Demissie Y, Mihret A, Abebe T. Bacterial profile and antimicrobial susceptibility pattern of isolates among burn patients at Yekatit 12 hospital burn center, Addis Ababa, Ethiopia. Ethiop J Health Sci. 2013;23:209–16.
Seni J, Najjuka CF, Kateete DP, Makobore P, Joloba ML, Kajumbula H, Kapesa A, Bwanga F. Antimicrobial resistance in hospitalized surgical patients: a silently emerging public health concern in Uganda. BMC research notes. 2013;6:298.
Fahmey SS. Early-onset sepsis in a neonatal intensive care unit in beni suef, Egypt: bacterial isolates and antibiotic resistance pattern. Korean J Pediatr. 2013;56:332–7.
Buhalata SN, Kwesigabo G, Sembuche S, Aboud S, Temu MM, Changalucha JM. Genital tract infections in women attending sexually transmitted infection clinics in Mwanza, north-west Tanzania. Southern African Journal of Epidemiology and Infection. 2013;28:48–54.
Githii S, Revathi G, Muigai A, Kariuki S. Carriage rate and serotypes of streptococcus pneumoniae amongst children in thika hospital, Kenya. African Journal of Laboratory Medicine. 2013;2
Raji MA, Jamal W, Ojemhen O, Rotimi VO. Point-surveillance of antibiotic resistance in Enterobacteriaceae isolates from patients in a Lagos teaching hospital, Nigeria. Journal of Infection and Public Health. 2013;6:431–7.
Omoregie R, Egbe CA, Dirisu J, Ogefere HO. Microbiology of neonatal septicemia in a tertiary hospital in Benin City, Nigeria. Biomarkers and Genomic Medicine. 2013;5:142–6.
Paglietti B, Falchi G, Mason P, Chitsatso O, Nair S, Gwanzura L, Uzzau S, Cappuccinelli P, Wain J, Rubino S. Diversity among human non-typhoidal salmonellae isolates from Zimbabwe. Trans R Soc Trop Med Hyg. 2013;107:487–92.
Page A-L, de Rekeneire N, Sayadi S, Aberrane S, Janssens A-C, Rieux C, Djibo A, Manuguerra J-C, Ducou-le-Pointe H, Grais RF, et al. Infections in children admitted with complicated severe acute malnutrition in Niger. PLoS One. 2013;8:e68699.
Oladeinde BH, Omoregie R, Olley M, Anunibe JA, Onifade AA. A 5 - year surveillance of wound infections at a rural tertiary hospital in Nigeria. Afr Health Sci. 2013;13(2):351–6.
Odongo CO, Anywar DA, Luryamamoi K, Odongo P. Antibiograms from community-acquired uropathogens in Gulu, northern Uganda--a cross-sectional study. BMC Infect Dis. 2013;13:193.
Chiedozie Kingsley O, Onwuezobe Ifeanyi A, Emmanuel Edet A, Chukwuemeka Smart O. Bacteriologic profile and antibiotics susceptibility pattern of suspected septicaemic patients in Uyo, Nigeria. Research Journal of Medical Sciences. 2013;7:35–9.
Esebelahie NO, Newton-Esebelahie FO, Omoregie R. Aerobic bacterial isolates from infected wounds. Afr J Clin Exp Microbiol. 2013;14:155–9.
Mshangila B, Paddy M, Kajumbula H, Ateenyi-Agaba C, Kahwa B, Seni J. External ocular surface bacterial isolates and their antimicrobial susceptibility patterns among pre-operative cataract patients at Mulago National Hospital in Kampala Uganda. BMC ophthalmology. 2013;13:71.
Meyer E, Whitelaw A, Edkins O, Fagan JJ. Chronic otorrhoea: spectrum of microorganisms and antibiotic sensitivity in a south African cohort. South African medical journal =. Suid-Afrikaanse tydskrif vir geneeskunde. 2013;103:471–3.
Bissong MEA, Fon PN, Tabe-Besong FO, Akenji TN. Asymptomatic bacteriuria in diabetes mellitus patients in Southwest Cameroon. Afr Health Sci. 2013;13:661–6.
Ibrahim ME, Bilal NE, Magzoub MA, Hamid ME. Prevalence of extended-spectrum β-lactamases-producing Escherichia Coli from hospitals in Khartoum state, Sudan. Oman Medical Journal. 2013;28:116–20.
Lunguya O, Lejon V, Phoba M-F, Bertrand S, Vanhoof R, Glupczynski Y, Verhaegen J, Muyembe-Tamfum J-J, Jacobs JO L et al: Antimicrobial Resistance in Invasive Non-typhoid Salmonella from the Democratic Republic of the Congo: Emergence of Decreased Fluoroquinolone Susceptibility and Extended-spectrum Beta Lactamases PLoS Neglected Tropical Diseases 2013, 7:e2103.
Lengerh A, Moges F, Unakal C, Anagaw B. Prevalence, associated risk factors and antimicrobial susceptibility pattern of Campylobacter species among under five diarrheic children at Gondar University Hospital, Northwest Ethiopia. BMC Pediatr. 2013;13:82.
Iregbu KC, Nwajiobi-Princewill PI. Urinary tract infections in a tertiary hospital in abuja, nigeria. Afr J Clin Exp Microbiol. 2013;14:169–73.
Fadeyibi IO, Raji MA, Ibrahim NA, Ugburo AO, Ademiluyi S. Bacteriology of infected burn wounds in the burn wards of a teaching hospital in Southwest Nigeria. Burns. 2013;39:168–73.
Hancali A, Ndowa F, Bellaji B, Bennani A, Kettani A, Charof R, El Aouad R, A. H, F. N, B. B et al: Antimicrobial resistance monitoring in Neisseria gonorrhoeae and strategic use of funds from the Global Fund to set up a systematic Moroccan gonococcal antimicrobial surveillance programme. Sexually Transmitted Infections 2013, 89:iv24-iv27.
Hailemariam M, Abebe T, Mihret A, Lambiyo T. Prevalence of Neisseria gonorrhea and their antimicrobial susceptibility patterns among symptomatic women attending gynecology outpatient Department in Hawassa referral hospital, Hawassa, Ethiopia. Ethiop J Health Sci. 2013;23:10–8.
Godebo G, Kibru G, Tassew H. Multidrug-resistant bacterial isolates in infected wounds at Jimma University Specialized Hospital, Ethiopia. Ann Clin Microbiol Antimicrob. 2013;12:17.
Schaumburg F, Alabi A, Kokou C, Grobusch MP, Köck R, Kaba H, Becker K, Adegnika AA, Kremsner PG, Peters G, et al. High burden of extended-spectrum β-lactamase-producing enterobacteriaceae in Gabon. J Antimicrob Chemother. 2013;68:2140–3.
Donkor ES, Foster-Nyarko E, Enweronu-Laryea CC. Relationship between antibiotic resistance and sickle cell anemia: preliminary evidence from a pediatric carriage study in Ghana. Infection and Drug Resistance. 2013;6:71–7.
Maina EK, Kiiyukia C, Wamae CN, Waiyaki PG, Kariuki S. Characterization of methicillin-resistant Staphylococcus Aureus from skin and soft tissue infections in patients in Nairobi, Kenya. Int J Infect Dis. 2013;17:e115–9.
Chika E, Ifeanyichukwu I, Michael A, Charles E. Susceptibility and detection of extended spectrum β-lactamase enzymes from otitis media pathogens. Am J Infect Dis. 2013;9:24–9.
Dinda V, Gunturu R, Kariuki S, Hakeem A, Raja A, Kimang'a A. Pattern of pathogens and their sensitivity isolated from surgical site infections at the Aga khan university hospital, Nairobi, Kenya. Ethiop J Health Sci. 2013;23:141–9.
Dayie NTKD, Arhin RE, Newman MJ, Dalsgaard A, Bisgaard M, Frimodt-Moller N, Slotved H-C. Penicillin resistance and serotype distribution of Streptococcus Pneumoniae in Ghanaian children less than six years of age. BMC Infect Dis. 2013;13:490.
Dagnew M, Yismaw G, Gizachew M, Gadisa A, Abebe T, Tadesse T, Alemu A, Mathewos B. Bacterial profile and antimicrobial susceptibility pattern in septicemia suspected patients attending Gondar University Hospital, Northwest Ethiopia. BMC research notes. 2013;6:283.
Coetzee E, Rode H, Kahn D. Pseudomonas Aeruginosa burn wound infection in a dedicated paediatric burns unit. South African journal of surgery Suid-Afrikaanse tydskrif vir chirurgie. 2013;51:50–3.
Christopher A, Mshana SE, Kidenya BR, Hokororo A, Morona D. Bacteremia and resistant gram-negative pathogens among under-fives in Tanzania. Ital J Pediatr. 2013;39:27.
Simango C. Antimicrobial susceptibility of campylobacter species. Southern African Journal of Epidemiology and Infection. 2013;28:139–42.
Bonkoungou IJO, Haukka K, Osterblad M, Hakanen AJ, Traore AS, Barro N, Siitonen A. Bacterial and viral etiology of childhood diarrhea in Ouagadougou Burkina Faso. BMC pediatrics. 2013;13:36.
Acquah SEK, Quaye L, Sagoe K, Ziem JB, Bromberger PI, Amponsem AA. Susceptibility of bacterial etiological agents to commonly-used antimicrobial agents in children with sepsis at the tamale teaching hospital. BMC Infect Dis. 2013;13:89.
Tsai AY, Dueger E, Macalino GE, Montano SM, Mbuchi M, Puplampu N, McClelland RS, Sanchez JL. Neisseria gonorrhoeae (GC) resistance surveillance in selected populations of five countries. Sex Transm Infect. 2013;89
Dau AA, Tloba S, Daw MA: Characterization of wound infections among patients injured during the 2011 Libyan conflict. East Mediterr Health J 2013, 19:356–361.
Adegoke AA, Komolafe AO, Aiyegoro OA. Rare bacterial meningitis among hospitalized children in a teaching hospital. Journal of Pure and Applied Microbiology. 2013;7:973–80.
Negussie A, Mulugeta G, Bedru A, Ali I, Shimeles D, Lema T, Aseffa A. Bacteriological profile and antimicrobial sensitivity pattern of blood culture isolates among septicemia-suspected children at Tikur Anbessa specialized hospital and Yekatit 12 hospital, Addis Ababa, Ethiopia. Crit Care. 2013;17
Shibabaw A, Abebe T, Mihret A. Antimicrobial susceptibility pattern of nasal Staphylococcus Aureus among Dessie referral hospital health care workers, Dessie, Northeast Ethiopia. Int J Infect Dis. 2014;25:e22–5.
Labi A-K, Obeng-Nkrumah N, Addison NO, Donkor ES. Salmonella blood stream infections in a tertiary care setting in Ghana. BMC Infect Dis. 2014;14:3857.
Smith AM, Mthanti MA, Haumann C, Tyalisi N, Boon GPG, Sooka A, Keddy KH. Nosocomial outbreak of salmonella enterica serovar typhimurium primarily affecting a pediatric ward in South Africa in 2012. J Clin Microbiol. 2014;52:627–31.
Abejew AA, Denboba AA, Mekonnen AG. Prevalence and antibiotic resistance pattern of urinary tract bacterial infections in Dessie area North-East Ethiopia. BMC research notes. 2014;7:687.
Aiken AM, Mutuku IM, Sabat AJ, Akkerboom V, Mwangi J, Scott JA, Morpeth SC, Friedrich AW, Grundmann H. Carriage of Staphylococcus Aureus in Thika level 5 hospital Kenya: a cross-sectional study. Antimicrob Resist Infect Control. 2014;3:22.
Akingbade O, Balogun S, Ojo D, Akinduti P, Okerentugba PO, Nwanze JC, Okonko IO, O. A, S. B, D. O et al: Resistant plasmid profile analysis of multidrug resistant Escherichia coli isolated from urinary tract infections in Abeokuta, Nigeria. Afr Health Sci 2014, 14:821–828.
Ako-Nai KA, Ebhodaghe BI, Osho P, Adejuyigbe E, Adeyemi FM, Kassim OO. Preponderance of bacterial isolates in urine of HIV-positive malaria-infected pregnant women with urinary tract infection. Journal of infection in developing countries. 2014;8:1591–600.
Ali MMM, Ahmed SF, Klena JD, Mohamed ZK, Moussa TAA, Ghenghesh KS. Enteroaggregative Escherichia Coli in diarrheic children in Egypt: molecular characterization and antimicrobial susceptibility. Journal of infection in developing countries. 2014;8:589–96.
Conceicao T, Santos Silva I, de Lencastre H, Aires-de-Sousa M: Staphylococcus aureus nasal carriage among patients and health care workers in Sao Tome and Principe. Microbial drug resistance (Larchmont, NY) 2014, 20:57–66.
Cornick JE, Harris SR, Parry CM, Moore MJ, Jassi C, Kamng'ona A, Kulohoma B, Heyderman RS, Bentley SD, Everett DB. Genomic identification of a novel co-trimoxazole resistance genotype and its prevalence amongst Streptococcus Pneumoniae in Malawi. J Antimicrob Chemother. 2014;69:368–74.
Dibua UM, Onyemerela IS, Nweze EI. Frequency, urinalysis and susceptibility profile of pathogens causing urinary tract infections in Enugu state, southeast Nigeria. Rev Inst Med Trop Sao Paulo. 2014;56(1):55–9.
Egyir B, Guardabassi L, Esson J, Nielsen SS, Newman MJ, Addo KK, Larsen AR, B. E, L. G, J. E et al: Insights into nasal carriage of Staphylococcus aureus in an urban and a rural community in Ghana. PLoS One 2014, 9:e96119.
Eleje GU, Adinma JI, Ghasi S, Ikechebelu JI, Igwegbe AO, Okonkwo JE, Okafor CI, Ezeama CO, Ezebialu IU, Ogbuagu CN, et al. Antibiotic susceptibility pattern of genital tract bacteria in pregnant women with preterm premature rupture of membranes in a resource-limited setting. Int J Gynecol Obstet. 2014;127:10–4.
Garedew-Kifelew L, Wondafrash N, Feleke A: Identification of drug-resistant Salmonella from food handlers at the University of Gondar, Ethiopia. BMC research notes 2014, 7:545.
Ibrahim ME, Bilal NE, Hamid ME. Comparison of phenotypic characteristics and antimicrobial resistance patterns of clinical Escherichia Coli collected from two unrelated geographical areas. Global journal of health science. 2014;6:126–35.
Isendahl J, Manjuba C, Rodrigues A, Xu W, Henriques-Normark B, Giske CG, Naucler P. Prevalence of community-acquired bacteraemia in Guinea-Bissau: an observational study. BMC Infect Dis. 2014;14:3859.
Vandepitte J, Hughes P, Matovu G, Bukenya J, Grosskurth H, Lewis DA. High prevalence of ciprofloxacin-resistant gonorrhea among female sex workers in Kampala, Uganda (2008-2009). Sex Transm Dis. 2014;41:233–7.
Moroh J-LA, Fleury Y, Tia H, Bahi C, Lietard C, Coroller L, Edoh V, Coulibaly A, Labia R, Leguerinel I. Diversity and antibiotic resistance of uropathogenic bacteria from Abidjan. Afr J Urol. 2014;20:18–24.
Ramos JM, Perez-Tanoira R, Garcia-Garcia C, Prieto-Perez L, Bellon MC, Mateos F, Tisisano G, Yohannes T, Reyes F, Gorgolas M. Leprosy ulcers in a rural hospital of Ethiopia: pattern of aerobic bacterial isolates and drug sensitivities. Ann Clin Microbiol Antimicrob. 2014;13:47.
Iyamba J-ML, Wambale JM, Lukukula CM, Takaisi-Kikuni NB. High prevalence of methicillin resistant staphylococci strains isolated from surgical site infections in Kinshasa. Pan African Medical Journal. 2014;18
Palle JN, Bassah N, Kamga HLF, Nkwelang G, Akoachere JF, Mbianda E, Nwarie UG, Njunda AL, Assob NJC, Ekane GH, et al. Current antibiotic susceptibility profile of the bacteria associated with surgical wound infections in the buea health district in Cameroon. Afr J Clin Exp Microbiol. 2014;15:27–34.
Kanj SS, Whitelaw A, Dowzicky MJ. In vitro activity of tigecycline and comparators against gram-positive and gram-negative isolates collected from the Middle East and Africa between 2004 and 2011. Int J Antimicrob Agents. 2014;43:170–8.
Kariuki S, Dougan G. Antibacterial resistance in sub-Saharan Africa: an underestimated emergency. Ann N Y Acad Sci. 2014;1323:43–55.
Kihla AJ-FT, Ngunde PJ, Evelyn MS, Gerard N, Ndip RN. Risk factors for wound infection in health care facilities in Buea Cameroon: Aerobic bacterial pathogens and antibiogram of isolates. Pan African Medical Journal. 2014;18:6.
Kouegnigan Rerambiah L, Ndong J-C, Mbakob Mengue Massoua P, Medzegue S, Elisee-Ndam M, Mintsa-Ndong A, Djoba Siawaya JF. Antimicrobial profiles of bacterial clinical isolates from the Gabonese National Laboratory of public health: data from routine activity. Int J Infect Dis. 2014;29:e48–53.
Kibret M, Abera B. Prevalence and antibiogram of bacterial isolates from urinary tract infections at Dessie Health Research Laboratory, Ethiopia. Asian Pacific Journal of Tropical Biomedicine. 2014;4:164–8.
Maltha J, Guiraud I, Kabore B, Lompo P, Ley B, Bottieau E, Van Geet C, Tinto H, Jacobs J. Frequency of severe malaria and invasive bacterial infections among children admitted to a rural hospital in Burkina Faso. PLoS One. 2014;9:e89103.
Mama M, Abdissa A, Sewunet T. Antimicrobial susceptibility pattern of bacterial isolates from wound infection and their sensitivity to alternative topical agents at Jimma University Specialized Hospital, South-West Ethiopia. Ann Clin Microbiol Antimicrob. 2014;13:14.
Manyahi J, Matee MI, Majigo M, Moyo S, Mshana SE, Lyamuya EF. Predominance of multi-drug resistant bacterial pathogens causing surgical site infections in Muhimbili National Hospital Tanzania. BMC research notes. 2014;7:500.
Moremi N, Mushi MF, Fidelis M, Chalya P, Mirambo M, Mshana SE. Predominance of multi-resistant gram-negative bacteria colonizing chronic lower limb ulcers (CLLUs) at Bugando medical center. BMC research notes. 2014;7:211.
Mukhtar A, Abdelaal A, Hussein M, Dabous H, Fawzy I, Obayah G, Hasanin A, Adel N, Ghaith D, Bahaa M, et al. Infection complications and pattern of bacterial resistance in living-donor liver transplantation: a multicenter epidemiologic study in Egypt. Transplant Proc. 2014;46:1444–7.
Mulatu G, Beyene G, Zeynudin A. Prevalence of Shigella, salmonella and campylobacter species and their susceptibility patters among under five children with diarrhea in Hawassa town, south Ethiopia. Ethiop J Health Sci. 2014;24:101–8.
Phoba M-F, De Boeck H, Ifeka BB, Dawili J, Lunguya O, Vanhoof R, Muyembe J-J, Van Geet C, Bertrand S, Jacobs J. Epidemic increase in salmonella bloodstream infection in children, Bwamanda, the Democratic Republic of Congo. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 2014;33:79–87.
Randrianirina F, Ratsima EH, Ramparany L, Randremanana R, Rakotonirina HC, Andriamanantena T, Rakotomanana F, Rajatonirina S, Richard V, Talarmin A. Antimicrobial resistance of bacterial enteropathogens isolated from stools in Madagascar. BMC Infect Dis. 2014;14:104.
Moyo SJ, Aboud S, Blomberg B, Mkopi N, Kasubi M, Manji K, Lyamuya EF, Maselle SY, Langeland N. High nasal carriage of methicillin-resistant staphylococcus aureus among healthy tanzanian under-5 children. Microb Drug Resist. 2014;20:82–8.
Scherbaum M, Kosters K, Murbeth RE, Ngoa UA, Kremsner PG, Lell B, Alabi A, M. S, K. K, R.E. M et al: Incidence, pathogens and resistance patterns of nosocomial infections at a rural hospital in Gabon. BMC Infect Dis 2014, 14:124.
Swann O, Everett DB, Furyk JS, Harrison EM, Msukwa MT, Heyderman RS, Molyneux EM. Bacterial meningitis in Malawian infants <2 months of age: etiology and susceptibility to World Health Organization first-line antibiotics. Pediatr Infect Dis J. 2014;33:560–5.
Vernet G, Mary C, Altmann DM, Doumbo O, Morpeth S, Bhutta ZA, Klugman KP. Surveillance for antimicrobial drug resistance in under-resourced countries. Emerg Infect Dis. 2014;20(3):434–41.
Wondimeneh Y, Muluye D, Alemu AA, Atinafu A, Yitayew G, Gebrecherkos T, Alemu AA, Damtie D, Ferede G: Urinary tract infection among obstetric fistula patients at Gondar University Hospital, Northwest Ethiopia. BMC Womens Health 2014, 14:12.
Adugna A, Kibret M, Abera B, Nibret E, Adal M. Antibiogram of e. Coli serotypes isolated from children aged under five with acute diarrhea in bahir dar town. Afr Health Sci. 2015;15:656–64.
Abujnah AA, Zorgani A, Sabri MAM, El-Mohammady H, Khalek RA, Ghenghesh KS. Multidrug resistance and extended-spectrum β-lactamases genes among Escherichia Coli from patients with urinary tract infections in northwestern Libya. Libyan Journal of Medicine. 2015;10
Wasihun AG, Wlekidan LN, Gebremariam SA, Welderufael AL, Muthupandian S, Haile TD, Dejene TA. Diagnosis and treatment of typhoid fever and associated prevailing drug resistance in northern Ethiopia. Int J Infect Dis. 2015;35:e96–e102.
Ernest AI, Ng’Walida N, Ndaboine E, Massinde A, Kihunrwa A, Mshana S. Maternal vaginorectal colonization by group B streptococcus and listeria monocytogenes and its risk factors among pregnant women attending tertiary hospital in Mwanza, Tanzania. Tanzania Journal of Health Research. 2015;17
Aamodt H, Mohn SC, Maselle S, Manji KP, Willems R, Jureen R, Langeland N, Blomberg B, H. A, S.C. M et al: Genetic relatedness and risk factor analysis of ampicillin-resistant and high-level gentamicin-resistant enterococci causing bloodstream infections in Tanzanian children. BMC Infect Dis 2015, 15:107.
Abou Shady HM, Bakr AE, Hashad ME, Alzohairy MA. Staphylococcus Aureus nasal carriage among outpatients attending primary health care centers: a comparative study of two cities in Saudi Arabia and Egypt. Braz J Infect Dis. 2015;19(1):68–76.
Alibi S, Ferjani A, Boukadida J. Molecular characterization of extended spectrum beta-lactamases produced by Klebsiella Pneumoniae clinical strains from a Tunisian hospital. Medecine et maladies infectieuses. 2015;45:139–43.
Amissah NA, Glasner C, Ablordey A, Tetteh CS, Kotey NK, Prah I, van der Werf TS, Rossen JW, van Dijl JM, Stienstra Y. Genetic diversity of Staphylococcus Aureus in Buruli ulcer. PLoS Negl Trop Dis. 2015;9:e0003421.
Anago E, Ayi-Fanou L, Akpovi CD, Hounkpe WB, Agassounon-Djikpo Tchibozo M, Bankole HS, Sanni A. Antibiotic resistance and genotype of beta-lactamase producing Escherichia Coli in nosocomial infections in Cotonou Benin. Annals of clinical microbiology and antimicrobials. 2015;14:5.
Ayepola OO, Olasupo NA, Egwari LO, Becker K, Schaumburg F. Molecular characterization and antimicrobial susceptibility of Staphylococcus Aureus isolates from clinical infection and asymptomatic carriers in Southwest Nigeria. PLoS One. 2015;10(9):e0137531.
Kwambana-Adams B, Darboe S, Nabwera H, Foster-Nyarko E, Ikumapayi UN, Secka O, Betts M, Bradbury R, Wegmüller R, Lawal B, et al. Salmonella infections in the Gambia, 2005-2015. Clin Infect Dis. 2015;61:S354–62.
Kraef C, Alabi AS, Peters G, Becker K, Kremsner PG, Rossatanga EG, Mellmann A, Grobusch MP, Zanger P, Schaumburg F. Co-detection of Panton-valentine leukocidin encoding genes and cotrimoxazole resistance in Staphylococcus Aureus in Gabon: implications for HIV-patients' care. Front Microbiol. 2015;6
Langendorf C, Le Hello S, Moumouni A, Gouali M, Mamaty A-A, Grais RF, Weill F-X, Page A-L. Enteric bacterial pathogens in children with diarrhea in niger: diversity and antimicrobial resistance. PLoS One. 2015;10:e0120275.
Mofolorunsho CK, Ocheni M, Omatola CA, Agieni AG. Staphylococcus Aureus prevalence and antibiotic susceptibility profile in anyigba, north-central Nigeria. Am J Infect Dis. 2015;11:93–7.
Shehab El-Din EMR, El-Sokkary MMA, Bassiouny MR, Hassan R. Epidemiology of neonatal sepsis and implicated pathogens: a study from Egypt. Biomed Res Int. 2015;2015
Nwankwo EO, Magaji NS, Tijjani J. Antibiotic susceptibility pattern of extended spectrum betalactamase (ESBL) producers and other bacterial pathogens in Kano, Nigeria. Trop J Pharm Res. 2015;14:1273–8.
Iliyasu G, Habib AG, Aminu MB. Antimicrobial susceptibility pattern of invasive pneumococcal isolates in north West Nigeria. J Global Infect Dis. 2015;7:70–4.
Tesfaw G, Kibru G, Mekonnen D, Abdissa A. Prevalence of group a β-haemolytic streptococcus among children with pharyngitis in Ethiopia. Eur Respir J. 2015;46
Hamdan HZ, Kubbara E, Adam AM, Hassan OS, Suliman SO, Adam I. Urinary tract infections and antimicrobial sensitivity among diabetic patients at Khartoum Sudan. Annals of clinical microbiology and antimicrobials. 2015;14:26.
Mandomando I, Bassat Q, Sigaúque B, Massora S, Quintó L, Ácacio S, Nhampossa T, Vubil D, Garrine M, Macete E, et al. Invasive salmonella infections among children from rural Mozambique, 2001-2014. Clin Infect Dis. 2015;61:S339–45.
Onwuezobe IA, Orok FE. Extended spectrum beta-lactamase producing uropathogens in asymptomatic pregnant women attending antenatal care in an urban community secondary health facility. Afr J Clin Exp Microbiol. 2015;16:49–53.
Iregbu KC, Eze SO. Pseudomonas Aeruginosa infections in a tertiary hospital in Nigeria. Afr J Clin Exp Microbiol. 2015;16:33–6.
Kityamuwesi R, Muwaz L, Kasangaki A, Kajumbula H, Rwenyonyi CM. Characteristics of pyogenic odontogenic infection in patients attending Mulago hospital Uganda: A cross-sectional study. BMC Microbiology. 2015;15:46.
Kalonji LM, Post A, Phoba M-F, Falay D, Ngbonda D, Muyembe J-J, Bertrand S, Ceyssens P-J, Mattheus W, Verhaegen J, et al. Invasive salmonella infections at multiple surveillance sites in the Democratic Republic of the Congo, 2011-2014. Clin Infect Dis. 2015;61:S346–53.
El Bouamri MC, Arsalane L, El Kamouni Y, Zouhair S. Antimicrobial susceptibility of urinary Klebsiella Pneumoniae and the emergence of carbapenem-resistant strains: a retrospective study from a university hospital in Morocco, North Africa. Afr J Urol. 2015;21:36–40.
Ntirenganya C, Manzi O, Muvunyi CM, Ogbuagu O. High prevalence of antimicrobial resistance among common bacterial isolates in a tertiary healthcare facility in Rwanda. Am J Trop Med Hyg. 2015;92:865–70.
Ochieng N, Okechi H, Ferson S, Albright AL. Bacteria causing ventriculoperitoneal shunt infections in a Kenyan population. J Neurosurg Pediatr. 2015;15:150–5.
Onken A, Said AK, Jorstad M, Jenum PA, Blomberg B. Prevalence and antimicrobial resistance of microbes causing bloodstream infections in Unguja Zanzibar. PLoS One. 2015;10:e0145632.
Peterside O, Pondei K, Akinbami FO. Bacteriological profile and antibiotic susceptibility pattern of neonatal sepsis at a teaching hospital in Bayelsa state, Nigeria. Tropical Medicine and Health. 2015;43:183–90.
Preziosi M, Zimba TF, Lee K, Tomas M, Kinlin S, Nhatave-Paiva C, Bene R, Paunde T, Lopes H, Kalkhoff S, et al. A prospective observational study of bacteraemia in adults admitted to an urban Mozambican hospital. S Afr Med J. 2015;105:370–4.
Eshetie S, Unakal C, Gelaw A, Ayelign B, Endris M, Moges F. Multidrug resistant and carbapenemase producing Enterobacteriaceae among patients with urinary tract infection at referral hospital, Northwest Ethiopia. Antimicrob Resist Infect Control. 2015;4
Saeed A, Abd H, Sandstrom G. Microbial aetiology of acute diarrhoea in children under five years of age in Khartoum, Sudan. J Med Microbiol. 2015;64:432–7.
Okoje-Adesomoju VN, Ifesanya JU, Alonge TO. Antimicrobial susceptibility pattern of oral microbial isolates among pregnant women in Ibadan south-east local government area, Nigeria. Afr J Biomed Res. 2015;18:29–35.
Mulu W, Yimer M, Zenebe Y, Abera B. Common causes of vaginal infections and antibiotic susceptibility of aerobic bacterial isolates in women of reproductive age attending at Felegehiwot referral hospital, Ethiopia: a cross sectional study. BMC Womens Health. 2015;15
Maina D, Omuse G, Revathi G, Adam RD. Spectrum of microbial diseases and resistance patterns at a private teaching Hospital in Kenya: implications for clinical practice. PLoS One. 2016;11:e0147659.
Odetoyin BW, Hofmann J, Aboderin AO, Okeke IN: Diarrhoeagenic Escherichia coli in mother-child Pairs in Ile-Ife, South Western Nigeria. BMC Infect Dis 2016.
Onanuga A, Selekere TL. Virulence and antimicrobial resistance of common urinary bacteria from asymptomatic students of Niger Delta University, Amassoma, Bayelsa state, Nigeria. Journal of Pharmacy and Bioallied Sciences. 2016;8:29–33.
Abdelaziz ZA, Ibrahim ME, Bilal NE, Hamid ME. Vaginal infections among pregnant women at Omdurman maternity Hospital in Khartoum, Sudan. Journal of Infection in Developing Countries. 2014;8:490–7.
Olorunmola FO, Kolawole DO, Lamikanra A. Antibiotic resistance and virulence properties in escherichia coli strains from cases of urinary tract infections. African Journal of Infectious Diseases. 2013;7:1–7.
Pondei K, Fente BG, Oladapo O. Current microbial isolates from wound swabs, their culture and sensitivity pattern at the Niger Delta University teaching hospital, Okolobiri, Nigeria. Tropical Medicine and Health. 2013;41:49–53.
Maina D, Makau P, Nyerere A, Revathi G. Antimicrobial resistance patterns in extended-spectrum β-lactamase producing Escherichia Coli and Klebsiella Pneumoniae isolates in a private tertiary hospital, Kenya. Microbiology Discovery. 2013;1
Sanford Guide - Trusted infectious disease recommendations: http://www.sanfordguide.com/stewardship/.
Acknowledgments
The authors would like to acknowledge Dr. Timothy Rodwell for commenting on the systematic review protocol as well as Piedra Lightfoot, Beatrice Gordis, Dr. Bill Rodriguez and Dr. Sophia Georghiou for critical review of the manuscript.
Ethics and consent to participate
Not applicable.
Funding
The Australian, UK, Dutch and Swiss Government and the WHO/TDR-program.
Availability of data and materials
Data supporting our findings can be found through the corresponding author (email: birknehtilahun@gmail.com) or the senior author (email: Sabine.Dittrich@finddx.org).
Author information
Authors and Affiliations
Contributions
BTT, IJG and SD conceived the idea, prepared the systematic review protocol and were involved in the literature search, screening and extraction of data. BTT performed the data analysis and developed the first draft of the manuscript. SD, IJG, SO, MW, JH, EA provided critical review of the manuscript and contributed to the final version. All the authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Detailed Methododologies (DOCX 24 kb)
Additional file 2:
Proportion of resistance of bacterial isolates to tested antibiotics by region and country (DOCX 263 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Tadesse, B.T., Ashley, E.A., Ongarello, S. et al. Antimicrobial resistance in Africa: a systematic review. BMC Infect Dis 17, 616 (2017). https://doi.org/10.1186/s12879-017-2713-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-017-2713-1