Abstract
Background
Subtribe Swertiinae, a medicinally significant and highly speciose Subtribe of family Gentianaceae. Despite previous extensive studies based on both morphology and molecular data, intergeneric and infrageneric relationships within subtribe Swertiinae remain controversial.
Methods
Here, we employed four newly generated Swertia chloroplast genomes with thirty other published genomes to elucidate their genomic characteristics.
Results
The 34 chloroplast genomes were small and ranged in size from 149,036 to 154,365 bp, each comprising two inverted repeat regions (size range 25,069–26,126 bp) that separated large single-copy (80,432–84,153 bp) and small single-copy (17,887–18,47 bp) regions, and all the chloroplast genomes showed similar gene orders, contents, and structures. These chloroplast genomes contained 129–134 genes each, including 84–89 protein-coding genes, 37 tRNAs, and 8 rRNAs. The chloroplast genomes of subtribe Swertiinae appeared to have lost some genes, such as rpl33, rpl2 and ycf15 genes. Comparative analyses revealed that two mutation hotspot regions (accD-psaI and ycf1) could serve as effective molecular markers for further phylogenetic analyses and species identification in subtribe Swertiinae. Positive selection analyses showed that two genes (ccsA and psbB) had high Ka/Ks ratios, indicating that chloroplast genes may have undergone positive selection in their evolutionary history. Phylogenetic analysis showed that the 34 subtribe Swertiinae species formed a monophyletic clade, with Veratrilla, Gentianopsis and Pterygocalyx located at the base of the phylogenetic tree. Some genera of this subtribe, however, were not monophyletic, including Swertia, Gentianopsis, Lomatogonium, Halenia, Veratrilla and Gentianopsis. In addition, our molecular phylogeny was consistent with taxonomic classification of subtribe Swertiinae in the Roate group and Tubular group. The results of molecular dating showed that the divergence between subtrib Gentianinae and subtrib Swertiinae was estimated to occur in 33.68 Ma. Roate group and Tubular group in subtribe Swertiinae approximately diverged in 25.17 Ma.
Conclusion
Overall, our study highlighted the taxonomic utility of chloroplast genomes in subtribe Swertiinae, and the genetic markers identified here will facilitate future studies on the evolution, conservation, population genetics, and phylogeography of subtribe Swertiinae species.
Similar content being viewed by others
Introduction
Subtribe Swertiinae, a medicinally significant and highly speciose Subtribe of family Gentianaceae, includes approximately 539–565 species, which are distributed in alpine, temperate and alpine regions around the world but rarely in tropical and subtropical regions at low latitudes. East Asia and North America are the centers of diversification of this subtribe, with 137 species of 11 genera in China [1]. Many species of subtribe Swertiinae, such as Halenia elliptica D.Don, Comastoma pedunculatum (Royle ex G.Don) Holub., Gentianopsis paludosa (Hook.f.) Ma., Lomatogonium carinthiacum (Wulfen) Rchb., Swertia mussotii Franch. and S. franchetiana Harry Sm., are the original traditional Tibetan folk medicinal species called “Dida” (Zangyinchen). These species have been used to cure bilious hepatitis, cholecystitis, hepatogenous jaundice, and hypoglycemia diseases in China for a long time [2, 3]. Therefore, increasing attention has been given to the plants of subtribe Swertiinae due to their extensive pharmacological effects.
A robust and well-resolved phylogeny in subtribe Swertiinae is fundamental to understanding the evolution and diversification of this subtribe, including individual traits, species diversification and conservation. Over the past decades, growing evidence of molecular markers (such as plastid markers and nuclear ribosomal DNA) has greatly advanced our understanding of subtribe Swertiinae relationships [4,5,6,7,8]. The results have shown that some genera of subtribe Swertiinae are not monophyletic groups, including Swertia, Gentianella, Comastoma, and Lomatogonium. In particular, Swertia is strongly paraphyletic to other genera. Meanwhile, the phylogenetic relationships between genera and within genera of subtribe Swertiinae obtained using different molecular markers (fewer fragments of chloroplast DNA sequences and ITS sequences) were also inconsistent [4,5,6,7,8]. For example, the molecular phylogeny of Swertiinae based on ITS and matK sequence data showed that the topmost part of the phylogenetic tree is a clade composed of Megacodon, Gentianella, Swertia, Jaeschke, Lomatogonium, and Comastoma, while the molecular phylogeny of Swertiinae based on matK and rbcL sequence data showed that the topmost part of the phylogenetic tree is a clade composed of some species of subgen. Ophelia of Swertia and, 7 species and varieties of the Gentianella. In addition, studies on anatomy [9,10,11], cytobiological chromosome numbers [12], pollen morphology [13], seed coat micromorphology [14,15,16] and other characteristics could not provide evidence for the definition of subtribe Swertiinae groups, showing that Swertia, Gentianella, and Lomatogonium are polyphyletic or paraphyletic genera. Therefore, effective methods for resolving phylogenetic relationships and evaluating previous classification of subtribe Swertiinae species are urgently needed.
Due to their moderate size, moderate nucleotide substitution rates and freedom from the problems of paralogy [17], plastid DNA sequences have been widely used for the reconstruction of plant phylogenies. A plastome phylogenomics approach has been successfully used to solve many enigmatic relationships within angiosperms and across all green plants [18,19,20,21,22,23,24]. Some studies have also revealed that chloroplast genome resources could offer helpful data for inferring the evolutionary relationships of Gentianaceae plants, thus reflecting vital evidence for a well-supported hypothesis of classification [25, 26]. To date, 34 complete chloroplast genomes of subtribe Swertiinae species have been sequenced, and some studies have been conducted based on these chloroplast genomes [25,26,27,28]. These studies initially probed the structural patterns of Swertia plastomes and the phylogenetic relationships within Swertia. Meanwhile, the phylogenetic relationships of subtribe Swertiinae were also examined. However, the phylogenetic relationships of subtribe Swertiinae obtained from the same chloroplast genome were inconsistent [25, 26]. Moreover, the structural patterns among genera within subtribe Swertiinae are still unknown. Thus, more evidence is crucial for further investigation.
The comparative analyses of the complete chloroplast genome structures among closely related species have yielded a wealth of information for taxonomically or phylogenetically recalcitrant lineages [29], such as Orychophragmus [30, 31], Epimedium [32], and Orchids [33]. In this study, we sequenced and reported the complete chloroplast genome of four Swertia species and conducted comparative chloroplast genomics with published chloroplast genomes of 30 other subtribe Swertiinae species. The study objectives were to (1) compare the sequences, structures, and gene organization of chloroplast genomes within subtribe Swertiinae; (2) identify polymorphic loci for future phylogenetic inference of the subtribe Swertiinae; and (3) elucidate the molecular evolution and phylogenetic relationships of subtribe Swertiinae.
Results
Structural features of subtribe Swertiinae chloroplast genomes
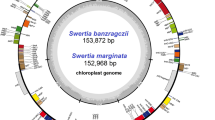
In this study, we analyzed the chloroplast genome features and gene contents of 34 species in 9 genera from subtribe Swertiinae (Table 1 and Table S1). All 34 chloroplast genomes displayed a typical quadripartite structure that was similar to that of the majority of angiosperm chloroplast genomes (Fig. 1). The length of the chloroplast genome of the 34 species varied between genera and species. The chloroplast genome length ranged from 149,036 (S. pubescens Franch.) to 154,365 bp (Pterygocalyx volubilis Maxim.), with an average length of 152,274 bp (Table 1). The longest chloroplast genome (154,365 bp) differed from the others by 0.614–5.329 kb. All complete chloroplast genomes were made up of four parts, namely, an LSC region (80,432–84,153 bp), an SSC region (17,887–18,476 bp), and two IR regions (25,069–26,126 bp). The GC content of the 34 species was very similar in both the whole chloroplast genome (37.5–38.26%) and the corresponding regions (LSC [32.18–36.36%], SSC [30.39%-33.66%], and IR [42.16%-43.38%]), with the IR regions having the highest GC contents (Table 1).
The chloroplast genome gene contents of the 34 species showed slight variation. The gene contents ranged from 129 (Gp. paludosa) to 134 (Gp. grandis (Harry Sm.) Ma, Veratrilla baillonii Franch, H. coreana S.M.Han, H.Won & C.E.Lim, S. bimaculata Hook.f. & Thomson ex C.B.Clarke, S. diluta (Turcz.) Zuev, S. leducii Franch., S. mussotii, S. souliei Burk., S. tetraptera Maxim., S. verticillifolia T.N.Ho & S.W.Liu and S. wolfgangiana Grüning) (Table 1). Accordingly, the number of protein-coding genes also varied, ranging from 84 to 89. However, the numbers of tRNA genes (37) and rRNA genes (8) were relatively conserved among species (Table S1). Among these protein-coding genes, four pseudogenes (rps16, infA, ycf1 and rps19) were found. Except for the lack of the rpl33 gene in the chloroplast genomes of S. dilatate C.B.Clarke, S. hispidicalyx Burkill, P. volubilis and C. pulmonarium (Turcz.) Toyokuni, the rpl2 gene in the chloroplast genome of C. falcatum (Turcz.) Toyokuni and the ycf15 gene in the chloroplast genome of G. paludosa, gene content differences were caused by the four pseudogenes. For example, due to the lack of the rps16, ycf1 and rps19 pseudogenes, the chloroplast genome of Lomatogoniopsis alpina T.N.Ho & S.W.Liu contained 131 genes (Table S1). Among all the genes, 18 genes (trnK-UUU, rps16, trnG-UCC, atpF, rpoC1, ycf3, trnL-UAA, trnV-UAC, rps12, clpP, petB, petD, rpl16, rpl2, ndhB, trnI-GAU, trnA-UGC and ndhA) in H. elliptica, V. baillonii and S. punicea Hemsl. contained only one intron, while 17 genes (rps16 was absent or did not contain an intron) in the remaining 31 species of subtribe Swertiinae contained one intron. Two protein-coding genes (ycf3 and clpP) in the chloroplast genomes of all 34 species contained two introns (Table S2).
The functions of major genes in the chloroplast genome of subtribe Swertiinae could be roughly divided into three categories (Table 2): photosynthesis, chloroplast self-replication and other. Genes associated with photosynthesis and self-replication made up the majority of the chloroplast genome.
SSR and codon usage analysis
The number of SSRs identified in the 34 subtribe Swertiinae chloroplast genomes ranged from 36 (S. bifolia Batalin and S. erythrosticta Maxim.) to 63 (S. cordata Wall.) (Fig. 2). Six types of repeat patterns were found among the SSRs, the numbers and types of which were different among the 34 chloroplast genomes. Among the mononucleotide repeats, A/T was dominant (50–82.22%), while C/G was rare (0–10.53%). Dinucleotides (1.89–11.63%), trinucleotides (4.35–19.44%) and pentanucleotides (3.92–20.00%) were found in all samples. Tetranucleotides and hexanucleotides were identified in eighteen and nine samples, respectively (Fig. 3 and Table S3).
Codon contents of 20 amino acids and stop codons in all protein-coding genes of four Subtribe Swertiinae species chloroplast genomes. The top panel shows the RSCU for the corresponding amino acids. The colored block represents different codons, which are shown in the panel below. (Note: 1 represents Halenia elliptica; 2 represents Swertia tetraptera; 3 represents S. mussotii; 4 represents Veratrilla baillonii; A represents Alanine; C represents Cysteine; D represents Aspartic acid; E represents Glutamic acid; F represents Phenylalanine; G represents Glycine; H represents Histidine; I represents Isoleucine; K represents Lysine; L represents Leucine; M represents Methionine; N represents Asparagine; P represents Proline; Q represents Glutamine; R represents Arginine; S represents Serine; U represents Threonine; V represents Valine; W represents Tryptophan; Y represents Tyrosine)
Codon usage frequency of the 34 subtribe Swertiinae chloroplast genomes was detected based on the sequences of protein-coding genes (CDSs). The number of codons of protein-coding genes in the 34 chloroplast genomes ranged from 20,531 (S. tetraptera) to 26,402 (H. elliptica). In all species, serine (Ser; 1075–2268 instances) was the most abundant amino acid encoded by four codons, followed by arginine (Arg; 1137–2244 instances), encoded by six codons (Table S4). In contrast, methionine and tryptophan were encoded by only one codon, with instances ranging from 219 to 610 and from 387–605, respectively, and showed no codon usage bias (RSCU = 1). The AGA codon for arginine had the largest RSCU values (1.70–2.11), and the CUG codon for leucine had the smallest RSCU values (0.31–0.80) in the 34 chloroplast genomes. A total of 26 codons with RSCU values greater than one were identified among the 64 codons in the chloroplast genomes. Twenty-three of the 26 codons with RSCU values greater than one ended with A or U, which showed the codon preferences in the chloroplast genomes (Fig. 3, Table S4).
GC content of 34 CDS sequences ranged from 0.384 to 0.402, while GC3s content of 34 CDS sequences was between 0.282 and 0.451(Table 3). The average content of T3s, C3s, A3s, G3s, GC3s and GC content of 34 CDS sequences were 0.4122, 0.2518, 0.3765, 0.2368, 0.372 and 0.397 respectively (Table 3). The average ENC value was 53.64, the minimum ENC value of S. franchetiana was 50.62, and the highest value of S. tetraptera was 57.76 (Table 3). All ENC values of this subtribe were greater than 49, reflecting weak codon bias overall. The CAI value of all species ranged from 0.149–0.164.
Comparative genomice analysis
We used the online tool mVISTA to identify the potential divergent sequences among the 34 subtribe Swertiinae chloroplast genomes, with the chloroplast genome of V. baillonii as a reference. The structures and sequences of the chloroplast genomes were conserved, especially in the IR regions (Fig. 4). Meanwhile, we used DNASP software to calculate the variation rate of coding and noncoding regions. The variation rates of noncoding regions were generally higher than those of coding regions (Fig. 5). The variation in noncoding region genes ranged from 11.11 to 99.28%, with an average of 63.98%, whereas the variation in coding region genes ranged from 5.78 to 88.97%, with an average of 25.39%. The variation rates of both coding regions and noncoding regions in the IR region were lower than those in other regions. Additionally, the noncoding intergenic regions were highly divergent, especially trnC-GCA-petN, trnS-GCU-trnR-UCU, ndhC-trnV-UAC, trnC-GCA-petN, psbM-trnD-GUC, trnG-GCC-trnfM-CAU, trnS-GGA-rps4, ndhC-trnV-UAC, accD-psaI, psbH-petB, rpl36-infA and rps15-ycf1. However, highly divergent regions were also found within protein-coding regions, such as in ycf3, petD, ndhF, petL, rpl20, rpl15 and ycf1. In addition, no genomic rearrangements were detected in the alignment analysis of 34 subtribe Swertiinae chloroplast genomes.
Comparison and analysis based on chloroplast genome of 34 Subtribe Swertiinae species. Orientation of genes were pointed out by arrows up the alignments. Purple, blue, pink and grey bars correspond to exons, untranslated regions, non-coding sequences and mRNA respectively. Y-axis indicates the genetic similarity percentage. Genetic similarity among 50-100% were showed in the figure. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Gene selective pressure analysis
We calculated the nonsynonymous (Ka) and synonymous (Ks) substitution ratios for 80 protein-coding genes to estimate the selection pressure on chloroplast genes by comparing L. alpina T.N.Ho & S.W.Liu with 33 other species in subtribe Swertiinae. The ratios of sixty-three protein coding genes could not be calculated because of Ka or Ks = 0, demonstrating that no synonymous or nonsynonymous changes occurred. For the remaining 17 protein-coding genes, the results indicated that the mean Ka/Ks ratio between L. alpina and the 33 other subtribe Swertiinae species ranged from 0.01 (rpl14) to 2.34 (psbB) (Fig. 6). However, the Ka/Ks ratio for most genes was less than one, showing that they underwent negative selection, except for ccsA and psbB, which experienced positive selection (Ka/Ks > 1).
Phylogenetic analysis
We used the complete chloroplast genome sequences and 80 shared protein sequences of 34 species from subtribe Swertiinae to construct phylogenetic trees with G. straminea, G. ovata and A. microlobus as outgroups. Phylogenetic trees built with the whole chloroplast genome and CDSs had the same topology (Figure S1). The Bayesian trees demonstrated that all species in the subtribe Swertiinae formed a monophyletic clade with high support in term of Bayesian posterior probabilities (PP = 1; Fig. 7). Additionally, this well-supported clade was divided into two major clades (A and B) within subtribe Swertiinae. Clade A was located at the base of the phylogenetic tree and was divided into two subclades (A1 and A2). The A1 subclade (P. volubilis) was sister to the A2 subclade consisting of three species of Gentianopsis and V. baillonii. Interestingly, G. paludosa did not cluster with the other two species of the same genus but clustered with V. baillonii, indicating that G. paludosa was closely related to V. baillonii. Clade B contained 29 species from the remaining 6 genera of subtribe Swertiinae, which formed three main branches in the phylogenetic tree (B1, B2 and B4), that is, the subgen. Swertia branch (B1), Gen. Halenia- Swertia dichotoma- Gen. Sinoswertia- Swertia bimaculate branch (B2) and subgen. Ophelia-Gen. Comastoma-L. alpina-Lomatogonium perenne T.N. Ho & S.W. Liu ex J. X. Yang branch (B4).
Phylogenetic tree of 34 Subtribe Swertiinae species using Bayesian inference (BI) analyses based on whole chloroplast genomes. Numbers at nodes correspond to Bayesian inference (BI) posterior probabilities (only probabilities ≥ 0.95 are shown). Different colors represent different clades: bule represents A1, red represents A2, purple represents B1, chartreuse represents B2, pink represents B3 and green represents B4
Divergence time estimate
In this study, complete chloroplast genomes sequences of 34 subtribe Swertiinae plants and three outgroups were used to estimate the divergence times of major clades in the subtribe Swertiinae. The divergence between subtrib Gentianinae and subtrib Swertiinae was estimated to occur in 33.68 Ma (Fig. 8). Roate group and Tubular group in Subtribe Swertiinae approximately diverged in 25.17 Ma. As far as the current 9 genera were concerned, Pterygocalyx was the oldest group of subtribe Swertiinae, diverged at 24.13 Ma. And then, Gentianopsis and Veratrilla diverged at 24.13 Ma. Swertia probably originated at 12.45 Ma. Comatoma, Lomatogoniopsis and Lomatogonium constituted the most evolved clades of subtribe Swertiinae, diverged probably at 7.56 Ma. The estimated divergence time in 23 species of Swertia was between 7.51 and 0.01 Ma.
IR contraction and expansion
We used the IRscope online website (https://irscope.shinyapps.io/irapp/) to visualize the differences in the four boundaries of the LSC, SSC, and IRs. Comparison of all subtribe Swertiinae plastomes with three outgroups uncovered relatively stable IRs, with little expansion or contraction (Fig. 9). In these 37 plastomes, the LSC-IRa borders were located in the rps19 gene with the exception of the LSC-IRa border of L. perenne, H. elliptica and G. ovata. In the outgroup G. ovata, the LSC-IRa border was located within the ndhB gene, while in L. perenne, the LSC-IRa border was located in the rpl22 gene, showing that the border had shifted by 59 bp. In H. elliptica, the LSC-IRa border was located within the rpl22 gene, which had undergone contraction. The boundary of SSC-IRa was positioned in the ndhF gene, ycf1 pseudogene and the intergenic spacer region between the ycf1 pseudogene and ndhF. The exact position of the SSC-IRb border had shifted by 10 bp in C. falcatum, 8 bp in S. cincta Burkill, 4 bp in S. mussotii, 9 bp in S. dichotoma L., Sp. Pl., 5 bp in S. przewalskii Pissjauk., 15 bp in S. erythrosticta, 10 bp in S. cordata and 3 bp in the outgroup A. microlobus. The SSC/IRa border in all subtribe Swertiinae plastomes was located within the ycf1 gene with a few exceptions, and the sequences demonstrated length variabilities among species. The IRa/LSC border in most species’ chloroplast genomes was located at the junction of the trnH gene and the rps19 pseudogene. In the L. perenne chloroplast genome, the trnH gene was far inside the LSC region, and the rps19 pseudogene was positioned at the IRa/LSC border. In V. baillonii, L. alpina, G. paludosa, Gp. Barbata (Froel.) Ma, C. pulmonarium, S. przewalskii, S. nervosa Wall., S. multicaulis D.Don and S. cordata chloroplast genomes, rps19 pseudogenes were lost, and the IRa/LSC border was positioned at the trnH gene.
Contraction and expansion of inverted repeats at the junction of chloroplast genome. JLB: LSC/IRb; JSB: IRb/SSC; JSA: SSC/IRa; JLA: IRa/LSC. Arrows illustrate the distance of genes from the junction site as shown for rpl22 at JLB and for trnH at JLA. The scale bar above some genes illustrates the number of base pairs that each gene occupies in specific regions of the chloroplast, e.g., the scale bar above ndhF represents the part of the gene located in the IRb region and the SSC region
Discussion
Organization and features of chloroplast genomes
Our study compared the features, content, and organization of the chloroplast genomes of 34 species in subtribe Swertiinae, demonstrating that all of them exhibited the typical quadripartite structure found in vascular plants [34,35,36]. The length of the chloroplast genomes of the 34 species varied from 149,036 (S. pubescens) to 154,365 bp (P. volubilis), implying that they are relatively conserved, revealing only minor differences that changed their sizes. Differences in chloroplast genome length within genera and families have previously been reported, such as in Swertia (Gentianaceae) [27], Notopterygium (Apiaceae) [37] and Rhodiola (Crassulaceae) [38], and in the subfamily Coryloideae of Betulaceae [39]. In this study, the differences in the chloroplast genome length of 34 species in 9 genera of subtribe Swertiinae were mainly caused by expansion and contraction of the IR region [40].
The chloroplast genomes had similar GC contents (37.5-38.26%), indicating high species similarity. The GC content in the IR region (43.39%) was higher than that in the other two regions (LSC, 35.92%; SSC, 31.88%), which may be related to the presence of four rRNA sequences in these regions, e.g., rrn16, rrn23, rrn4.5, and rrn5, as previously reported in many complete chloroplast genomes of angiosperms [41].
Regarding gene estimates, we found some differences among the chloroplast genomes. Gene numbers ranged from 129 (Gp. paludosa) to 134 (Gp. grandis, V. baillonii, H. coreana, S. bimaculate, S. diluta, S. leducii, S. mussotii, S. souliei, S. tetraptera, S. verticillifolia and S. wolfgangiana). Gp. paludosa had 129 genes due to the absence of the ycf15 gene and pseudogenes rps16, rps19, infA and ycf1, while Gp. grandis, H. coreana, S. bimaculate, S. diluta, S. leducii, S. mussotii, S. souliei, S. tetraptera, S. verticillifolia and S. wolfgangiana contained 134 genes because of a duplication of rps19 and ycf1. In fact, duplicated rps19 and ycf1 pseudogenes have also been reported in other Gentiaceae species [42]. Similarly, rpl33 was lost from S. dilatate, S. hispidicalyx, P. volubilis and C. pulmonarium. According to relevant research on rpl33, this gene could maintain enough plastid translation ability in cold environments [43]. In the light of our results and previous studies, rpl33 exists in most of Gentianaceae members with wide distribution and large numbers of species. In contrast, rpl33 is not as common in less specious, like Gentianella and Pterygocalyx. Therefore, the gene rpl33 may confer greater tolerance of cold climates; this explains the relatively smaller number of Gentianella and Pterygocalyx on QTP. Additionally, there have been reports of the absence of ndh genes in other Gentiaceae species, including ndhA, ndhC, ndhG, ndhH, ndhI, ndhJ, and ndhK [44]. However, the lack of the ycf15 gene has not been reported. Thus, small changes in the content of these genes in the chloroplast genomes of subtribe Swertiinae are caused by evolutionary events of gene deletion and insertion.
Simple sequence repeats (SSRs) and codon usage analysis
Chloroplast SSRs usually show a high level of variation and are widely used in the study of polymorphism, population genetics and phylogenetics [29, 45, 46]. Our study analyzed the number of different SSR motifs in the chloroplast genomes of 34 subtribe Swertiinae species. Compared with that in other angiosperms, the number of chloroplast genome SSRs (36–63) in the 34 subtribe Swertiinae species was low to medium. Among the SSRs, a large number of single nucleotide repeats were detected, among which polyA and polyT structures were common, which was consistent with the results of previous studies [47,48,49,50]. These SSRs may be useful for subsequent analyses of interspecies genomic polymorphism and population genetics based on repeat length polymorphism. Experts have different views of the mechanisms explaining most SSRs in chloroplast genomes. Slip chain mismatch and intramolecular recombination are currently considered the main mechanisms that cause most SSRs [51].
The choice of synonymous codons for amino acids encoded by an organism’s genes is not completely random, and there is codon usage bias. Codon usage bias not only plays an important regulatory role in gene expression level but also helps to improve the accuracy and efficiency of translation. In addition to being affected by selection and mutation, codon usage is affected by tRNA abundance, base composition, gene position on chromosomes, gene length and expression level, amino acid hydrophobicity and mRNA secondary structure.
Previous studies have shown that analysis of codon bias in the chloroplast genome is helpful for understanding the origin and evolution of species [52]. In addition, the frequency of codon use is also related to gene expression. Nucleotide composition is one of the important factors affecting codon use bias. In the genome, the AT and GC contents are closely related to synonymous codon use bias. GC refers to the total content of all codons G and C, and GC3s pertains to the frequency of G and C bases in the third codon base of synonymous codons encoding the same amino acid. GC reflects the strength of directional mutation pressure, and GC3s is closely related to codon bias. The main difference in synonymous codons is reflected in the third base, and a change in this base of codons usually does not cause changes in encoded amino acids. Therefore, the selection pressure on the third base of the codon is less selective [53]. GC3s is used as an important basis for analyzing codon usage pattern [54]. GC and GC3s in the codons of these 34 chloroplast genomes were all less than 0.5, indicating that the chloroplast genomes of Subtribe Swertiinae species tended to use A/T bases and A/T-ending codons. Moreover, most amino acids in the 34 species chloroplast genomes had codon bias with a high preference (RSCU > 1), except for methionine and tryptophan (RSCU = 1). The RSCU value of codon types ending with A or U was larger than that for types ending with G or C, which showed a codon preference for bases A or U in the 34 species’ chloroplast genomes. Similar conclusions have been made in studies of Phyllanthaceae [55], Notopterygium [37], Cinnamomum camphora [56] and others. Codon adaptation index values and effective number of codon values indicated a slight bias in codon usage in the Subtribe Swertiinae species. CAI and ENC are important indexes to judge strength of codon bias [57]. In this study, CAI ranged from 0.149 to 0.164 in chloroplast genomes of 34 Subtribe Swertiinae species, with an average of 0.162, while ENc ranged from 50.62 to 57.76 in chloroplast genomes of 34 Subtribe Swertiinae species, with an average of 53.64. These results indicated that codon bias was weak in chloroplast genomes of Subtribe Swertiinae species, which might be related to the conserved of chloroplast genomes of Subtribe Swertiinae species [57]. In all, these findings advance our understanding of the evolutionary history of subtribe Swertiinae, especially in relation to natural selection and mutation pressures.
Comparative genomes and characterization of substitution rates
Although the chloroplast genome is considered to be fairly conserved in angiosperms, mutational hotspots are often found in the sequences of some closely related species. These mutational hotspots are widely used in plant phylogenetics, group genetics and DNA barcode research. Based on the nucleotide diversity analyses, we identified nineteen highly variable regions with high variation rates that can be used as markers to resolve taxonomic issues in subtribe Swertiinae and function as DNA barcodes for species identification.
The intergenic regions ndhC-trnV-UAC, trnC-GCA-petN, trnS-GCU-trnR-UCU, ndhC-trnV-UAC, trnC-GCA-petN, psbM-trnD-GUC, trnG-GCC-trnfM-CAU, trnS-GGA-rps4, and ndhC-trnV-UAC possess high variation rates (Fig. 5), however, these markers are rarely used in plant phylogenetics. The intergenic region accD-psaI is approximately 640 bp long and is used for research in Gentiana sect. Cruciata (Gentianaceae) [58], Pyrus (Rosaceae) [59], and Coryloideae (Betulaceae) [60]. Compared with existing candidate genes (matK and rbcL combination), the ycf1 gene is more suitable for barcodes of land plants due to its more variable loci [61]. Recently, ycf1 gene has been treated as the core DNA barcode in sect. Cruciata and found to provide sufficient genetic markers for resolving the phylogeny of Gentianaceae [58]. Based on our study, these two divergent markers may be useful for further phylogenetics and species identification of subtribe Swertiinae.
Phylogenetic and divergence time of Subtribe Swertiinae
The topologies of the ML and BI trees constructed with complete chloroplast genome sequences and shared protein-coding gene sequences were consistent, indicating that all 34 subtribe Swertiinae species formed a monophyletic clade, which was sister to Subtribe Gentianaceae. The monophyly of subtribe Swertiinae is therefore verified by chloroplast genome data, a finding consistent with that of previous studies [4,5,6,7,8]. Meanwhile, our molecular phylogeny agreed with the taxonomic classification of subtribe Swertiinae in the Roate group and Tubular group. Previous studies have shown that the Roate group contains Swertia, Lomatogonium, Lomatogoniopsis and Sinoswertia, while the Tubular group includes the rest. In this study, the well-supported clade was divided into two major clades (A and B) within subtribe Swertiinae, of which clade B represented the Roate group (except for Halenia) and clade A represented the Tubular group. P. volubilis was closely related to Gentianopsis and V. baillonii, which are located at the base of subtribe Swertiinae. In the other studies, the base groups of subtribe Swertiinae also included Obliaria, Latouchea, Bartonia and Megacodon. From the analysis of geographical distribution, the basal groups are mostly isolated monospecies or small genera containing only a few species, such as Obliaria (1 species) and Bartonia (4 species), distributed in North America. Latouchea (1 species) and Megacodon (2 species) are distributed in Southwest China and the Himalaya region. Pterygocalyx (1 species) is distributed in Asia, and Veratrilla (2 species) is distributed in southwest China, northeast India, Sikkim and Bhutan. From the perspective of morphology, except for Bartonia (no floral nectary was observed), Obliaria, Megacodon, Latouchea, Gentianopsis and Pterygocalyx all have floral nectaries at the base of the ovary, which is the same as Gentian of the outgroup and different from other genera of subtribe Swertiinae (most species have floral nectaries on corolla lobes). Thus, nectaries at the base of the ovary may be ancestral characteristics of subtribe Swertiinae. However, the position of V. baillonii is still unresolved because the results of this study were inconsistent with those of previous studies [25, 26, 28]. C. falcatum and C. pulmonstium formed a clade, which showed that Comastoma was as one monophyletic group. Our result supports previous studies based on ITS sequences and embryological characteristics [12, 62]. Comastoma clustered with L. perenne and L. alpina in a more recently evolved branch. The genus Halenia is closely related to S. tetraptera, S. dichotoma and S. bimaculata, which is consistent with the results of molecular systematics study based on ITS fragments by von Hagen and Kadereit (2002) [9] and embryology study by Xue et al. (2007) [63]. In addition, Halenia, Lomatogonium, Lomatogoniopsis, and Sinoswertia were polyphyletic with the closely related ally Swertia, whereas Veratrilla was polyphyletic with its closely related ally Gentianopsis, which indicated that these genera were not monophyletic groups. The molecular data of these genera conflicted with their morphological classification, which was consistent with the findings of previous studies. The inconsistencies may be explained by the fact that the morphological classification does not reflect the true evolutionary history. The most significant reason is the plasticity of morphological characteristics.
Exploitation and habitat destruction have seriously threatened the resources of famous Tibetan folk medicinal species. Phylogenetic relationships have substantial scientific significance for the sustainable use of rare resources of alpine medicinal plants. For example, the resources available for S. mussotii, the original traditional Tibetan folk medicinal species called “Dida”, cannot not meet the increasing market demand. An in-depth study on the genetic relationships of medicinal plants in subtribe Swertiinae showed that S. franchetiana, S. punicea and S. kouitchensis were closely related to S. mussotii, which has guiding significance for expanding drug sources.
Phylogenetic and divergence time analysis indicated that the subtribe Swertiinae species may have undergone rapid radiation. The divergence time analysis results indicated that subtribe Swertiinae originated in the end of the Eocene (Fig. 9). Two significant periods of rapid diversification of subtribe Swertiinae were found. The first was in the early Miocene. During this period, the QTP was further uplifted, and the Himalayan mountains and Tianshan Mountains were significantly elevated, which strongly changed the atmospheric circulation. Meanwhile, the global temperature decreased from the optimum temperature in the middle Miocene of the third century, resulting in a cool and dry climate [64,65,66,67]. The cool and dry climate resulted in suitable habitats for subtribe Swertiinae fragmentation, which led to the first rapid radiation and.led to the major lineages of the extant subtribe Swertiinae species. The second period was in the Quaternary, which led to most of the extant subtribe Swertiinae species. After 5 Ma, the global temperature decreased sharply after a short period of global warming [68], providing a diverse range of habitats and further increasing the species diversity of subtribe Swertiinae.
Adaptative evolution of subtribe Swertiinae
Synonymous and nonsynonymous nucleotide substitution patterns play a major role in adaptive evolution. In subtribe Swertiinae, we did not detect significant positive selection for the majority of genes, with only two genes (ccsA and psbB) showing possible positive selection; these may have played a vital role in adaptive evolution in subtribe Swertiinae. Our results were in accordance with those of a previous study, which showed that ccsA was under positive selection in the chloroplast genome of 15 angiosperms [69]. psbB, encoding photosystem subunits (Table 2), plays a vital role in the life history of plants. In addition, the ccsA gene is a c-type cytochrome synthesis gene (Table 2) in plants. The cssA gene is responsible for encoding the cytochrome c synthesis protein, which has approximately 250 ~ 350 amino acids and is a membrane binding protein. The coding product of ccsA can co-form the ccsA complex with the coding protein of another gene, ccsB [70]. Xie et al. (1996) [71] believed that the ccsA gene was related to the binding of cytochrome C-heme. This has implications for understanding the adaptive evolution of ccsA genes in angiosperms. These genes are highly correlated with physiological processes such as photosynthesis; thus, their positive selection on them may help subtribe Swertiinae species quickly adapt to all kinds of environments and enable their wide global distribution.
Plant species in subtribe Swertiinae are diverse and have important medicinal value. The current analysis of the chloroplast genome has revealed its unique sequence structure and rich genetic information. However, for the whole subtribe, the use of only 34 genome sequences is far from sufficient. In the next step, the genetic data of other taxa of this subtribe should be supplemented to further explore the phylogenetic relationships of this subtribe and provide a scientific basis for the conservation of species resources of this subtribe and the study of Chinese traditional medicine pharmacognosy.
Single-copy nuclear genes are helpful for elucidating the complex phylogenetic relationships of subtribe Swertiinae. Single-copy nuclear genes play an important role in molecular phylogenetics [72]. Compared with common nuclear genes, single-copy nuclear genes can reduce the influence of paralogous genes. Because of the large number of duplicate genes in the plant genome, it is difficult to screen single-copy nuclear genes [73]. However, with the development of high-throughput sequencing technology, it has been possible to develop a large number of single-copy nuclear genes at the whole-genome level [74, 75], and they have been applied in phylogenetic studies of several taxa [76]. However, no single-copy nuclear genes have been studied in subtribe Swertiinae. There are still many problems in the classification of subtribe Swertiinae, and the monophyly of several genera has not been determined, which reflects the complex evolutionary process of this Subtribe. Therefore, a large number of single-copy nuclear genes should be widely used to resolve the phylogenetic relationships of rapidly differentiated taxa such as those belonging to subtribe Swertiinae. Because the phylogenetic trees constructed from chloroplast data and nuclear gene data are often inconsistent [5], integrative analysis of the two is particularly important. At the same time, population genetics can be used to analyze the phylogenetic relationships and evolutionary processes of closely related taxa within genera or groups.
Conclusion
We presented a comparative analysis of 34 plastomes from 34 subtribe Swertiinae species and reported a comprehensive study of their phylogenetic relationships, divergence time estimates, and adaptative evolution. Phylogenetic analysis showed that the 34 species formed a monophyletic clade, and Veratrilla, Gentianopsis and Pterygocalyx were located at the base of the phylogenetic tree. In addition, our molecular phylogenetic results agreed with the taxonomic classification of subtribe Swertiinae in the Roate group and Tubular group. Positive selection analyses showed that two genes (ccsA and psbB) had high Ka/Ks ratios, indicating that chloroplast genes may have undergone positive selection during the evolutionary history of this group. Divergence time analysis revealed that subtribe Swertiinae originated in the end of the Eocene and diversified in the early Miocene Tertiary. Overall, the whole chloroplast genome sequences provide valuable information for elucidating the phylogeny, divergence time and evolutionary process of subtribe Swertiinae.
Materials and methods
Sampling
We collected fresh young leaves of S. tetraptera, S. franchetiana, S. przewalskii and S. bifolia. from Mengyuan County of Qinghai Province (101.32′E, 37.62′N, 3,208 m), Huangzhong County of Qinghai Province (101.63′E, 36.57′N, 2,510 m), Qilian County of Qinghai Province (99.61′E, 38.83′N, 3,234 m), and Qilian County of Qinghai Province (102.22′E, 37.45′N, 3,135 m), respectively. We used silica gel to rapidly store the leaves until dried. These samples were all identified by taxonomist Prof. Yuhu Wu at the Northwest Institute of Plateau Biology, Chinese Academy of Sciences. Voucher specimens of these four species were deposited in the Qinghai-Tibetan Plateau Museum of Biology (QTPMB) with voucher numbers QHGC-2011, QHGC20190821, QHGC-2013, and QHGC-2014, respectively. All of the published complete chloroplast genomes of Subtribe Swertiinae were downloaded from GenBank (Table S5). In total, 34 plastid genomes from 9 genera of subtribe Swertiinae species were obtained. Our experimental research, including the collection of plant materials, complies with institutional, national, or international guidelines.
DNA extraction, library preparation and genome sequencing
Total genomic DNA was extracted from dried leaves of the four Swertia L. plants by using an improved CTAB method [77] and estimated for purity and concentration using a NanoDrop 2000 microspectrophotometer. Each genomic DNA sample was broken into fragments of different lengths by ultrasound. Then, the DNA fragments were purified, the ends were repaired, an A tail was added to the 3' end, and the sequencing joints were connected. After that, agarose gel electrophoresis was used to select suitably sized DNA fragments, and PCR amplification was performed to complete the preparation of the sequencing library. After qualified library quality inspection, the Illumina HiSeq platform (Beijing Biomarker Technologies Co., Ltd.) was used for 150 bp paired-end sequencing.
Chloroplast genome assembly and annotation
Raw sequencing data were transformed into sequenced reads (raw data) by performing a base calling analysis of the raw image files. SQCToolkit_v2.3.3 software [78] was used to filter the raw read data obtained by sequencing to remove low-quality regions and obtain clean reads. The results were then stored in the FASTQ format. We used the iterative organelle genome assembly pipeline to assemble the chloroplast genome with S. mussotii (NC_031155) serving as a reference [79]. Then, SPAdes v3.6.1 software was employed for de novo splicing under default parameters and to generate a series of contigs [80]. Contigs larger than 1,000 bp were used for chloroplast genome assembly. Complete chloroplast genome sequences were constructed by matching and linking contigs [81] and filling the gaps after assembly using second-generation sequencing technology.
The chloroplast genomes of the four Swertia L. species were annotated using the online program GeSeq [82] and PGA software [83]. We compared annotations from the two methods and made final adjustments manually in Geneious version 11.0.2 [81]. Then, we checked the initial annotation and putative starts, stops, and intron positions by comparison with homologous genes in congeneric species S. mussotii. Then, we used OGDRAW [84] software to draw circular plastid genome maps of the four Swertia L. species. Finally, the sequence data and gene annotation information of the four Swertia L. species were uploaded to the NCBI database with accession numbers NC_056357 (S. franchetiana), ON164641 (S. przewalskii), ON017794 (S. tetraptera), and SUB11740174 (S. bifolia).
Sequence alignment
Including the 30 previously sequenced plastomes in GenBank, plastome sequences (using both coding and noncoding regions) from 34 species in Subtribe Swertiinae were aligned with MAFFT (version 7) [85] using the default settings. The plastomes of subtribe Swertiinae are conserved in gene order and content, so the alignment was straightforward; some poorly aligned regions were manually adjusted in GENEIOUSv.8.1 [81]. Different methods for handling gaps have a certain impact on the final results [86]. Therefore, it is very important to address the gaps in the data set when data are analyzed. As shown by previous research results, reliable gaps are within comparison areas of high confidence, showing a kind of indel event during the sequence evolution process. Therefore, reliable gaps also reflect evolutionary history of sequences, and should be treated as characters, just as bases. Unreliable gaps are in fuzzy comparison areas and categorized as sites of uncertain alignment. They are excluded from systematic analysis and not included in the analyzed data set. In this research, unreliable gaps were deleted from the compared sequences before analysis.
Single sequence repeat (SSR) and relative synonymous sodon usage (RSCU) analysis
We used the online MISA program [87] to detect SSRs in the chloroplast genomes of 34 species in subtribe Swertiinae using the following parameters: mononucleotide unit repetition number ≥ 10; dinucleotide unit repetition number ≥ 5; trinucleotide unit repetition number ≥ 4; and tetranucleotide, pentanucleotide, and hexanucleotide unit repetition number ≥ 3 [87]. CodonW1.4.2 software was also employed to confirm the amino acid usage frequency and relative synonymous codon usage (RSCU) [88].
Comparative analysis of complete chloroplast genome
We used IRscope software to visually analyze boundaries between the four main chloroplast regions (LSC/IRb/SSC/IRa) of 34 species in subtribe Swertiinae [89]. Moreover, Mauve software was used to analyze the chloroplast DNA rearrangement of the 34 species in subtribe Swertiinae. Meanwhile, the online software mVISTA was used to compare the 34 species of Subtribe Swertiinae in shuffle-LAGAN Mode [90]. The genome of V. Baillonii was used as the reference genome. The method developed by Zhang et al. (2011) [91] was used to calculate the percentages of variable characters in the coding and noncoding regions of the chloroplast genomes. The proportion of mutation events = [(NS + ID)/L] × 100, where NS = the number of nucleotide substitutions, ID = the number of indels, and L = the aligned sequence length.
Analysis of synonymous (Ks) and non-synonymous (Ka) substitution rate
We computed the selective pressures for protein-encoding genes that were located in three regions of the chloroplast genomes (LSC, SSC and one IR). Protein-coding genes that were shared by the 34 species were chosen and extracted from complete chloroplast genomes for synonymous (Ks) and nonsynonymous (Ka) substitution rate analysis. Each gene selection was forecast by taking into account the ratios of Ka/Ks, that is, Ka/Ks < 1 purifying selection, Ka/Ks = 1 neutral selection, and Ka/Ks > 1 positive selection [92]. Nonsynonymous (Ka) and synonymous (Ks) substitution rates were calculated using KaKs_Calculator 2.0 software [93] with the following settings: genetic code table 11 (bacterial and plant plastid code) and method of calculation: NG.
Phylogenetic analysis
To examine the phylogenetic relationship of 34 species of 9 genera within subtribe Swertiinae, an evolutionary tree was constructed with the complete chloroplast DNA sequences using G. straminea Maxim. (KJ657732), Gardneria ovata Wall. (NC_065470) and Amalocalyx microlobus Pierre (NC_067035) as outgroups. Meanwhile, we used 80 shared protein-coding genes of 34 chloroplast genomes to construct a molecular phylogenetic tree. Phylogenetic analyses were performed according to the Bayesian inference (BI) method under the best-fit substitution model GTR + I + G selected by the AIC in MrModeltest 2.3 [94] using MrBayes v3.2.1 [95]. BI analysis was run independently using four Markov Chain Monte Carlo (MCMC) chains, that is, three heated chains and one cold chain, and started with a random tree; each chain was run for 2 × 107 generations and sampled every 2 000 generations, and discarding the first 25% preheated trees as burn-in. We estimated the convergence of data runs using an average standard deviation of split frequencies (ASDSF) < 0.01 and Tracer v1.7.1 [96] to check for an effective sample size (ESS) > 200. The phylogenetic tree nodes were considered well-supported when the node Bayesian posterior probability (BP) was ≥ 0.95.
Divergence-time estimation
The divergence times of subtribe Swertiinae species was estimated using BEAST v1.8.4 software package [97]. The chloroplast genomes sequences from 34 species of subtribe Swertiinae and outgroups were included in the analysis. Firstly, BEAUti in the software package was used to set the parameters of nexus format sequence file. The optimal nucleotide substitution model selected by MrModeltest was GTR + I + G. An uncorrelated relaxed clock analysis with a Yule process speciation model was specified. Pollen of Lisianthius P. Browne (Trib. Potalieae, Gentianaceae) from Eocene sediments in Panama was used as lognormal priors, with an offset at 33.6 Ma [98], a mean of 0.7, and a standard deviation of 1.0, as used by Matuszak et al. (2015) [99]. The final Markov chain was run seven for 10 million generations, sampling every 10,00 generations. After the parameters were set, the BEAST file in xml format was generated. Secondly, the XML format file was imported into the BEAST software. Thirdly, Tracer v1.7.1 [96] was used to assess the convergence and stability of run parameters, i.e., ESS of parameters > 200. Fourthly, Maximum clade credibility (MCC) trees were produced using TreeAnnotator v2.4.1 [100] by applying a burn-in (as trees) of 10%, posterior probability limit of 0.5, and median height for node selection. Finally, FigTree v1.4.4 [101] was used to edit and visualize the time tree.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files. The datasets generated and/or analyzed during the current study are available in the GenBank repository (S. przewalskii: https://www.ncbi.nlm.nih.gov/nuccore/ON164641; S. franchetiana: https://www.ncbi.nlm.nih.gov/nuccore/NC_056357.1/; S. tetraptera: https://www.ncbi.nlm.nih.gov/nuccore/ON017794; S. bifolia: SUB11740174).
References
Struwe L, Albert VA. Gentianaceae: systematics and natural history. New York: Cambridge University Press; 2002. p. 242.
Fan G, Luo WZ, Luo SH, Li Y, Meng XL, Zhou XD, Zhang Y. Metabolic discrimination of Swertia mussotii and Swertia chirayita known as “Zangyinchen” in traditional Tibetan medicine by 1H NMR-based metabolomics. J Pharmaceut Biomed. 2007;98:364–70.
Wen J, Zhang Y, Liu C, Li XH, Fan G, Geng ZJ, Gu R. Research on origin, property and efficacy of Tibetan medicine “Dida.” Chin J Chinese Materia Medica. 2016;41(4):609–12.
Yuan YM, Küpfer P. Molecular phylogenetics of the subtribe Gentianinae (Gentianaceae) inferred from the sequences of internal transcribed spacers (ITS) of nuclear ribosomal DNA. Plant Syst Evol. 1995;196(3–4):207–26.
Chassot P, Nemomissa S, Yuan YM, Küpfer P. High paraphyly of Swertia L. (Gentianaceae) in the Gentianella-lineage as revealed by nuclear and chloroplast DNA sequence variation. Plant Syst Evol. 2001;229:1–21.
Liu JQ, Chen ZD, Lu AM. A preliminary analysis of the phylogeny of the Swertiinae (Gentianaceae) based on ITS data. Isr J Plant Sci. 2001;49(4):301–8.
Cao Q, Xu LH, Wang JL, Zhang FQ, Chen SL. Molecular phylogeny of subtribe Swertiinae. Bull Bot Res. 2021;41(3):408–18.
Xi HC, Sun Y, Xue CY. Molecular phylogeny of Swertiinae (Gentianaceae-Gentianeae) based on sequence data of ITS and matK. Plant Divers Resour. 2014;36(2):145–56.
von Hagen KB, Kadereit JW. Phylogeny and flower evolution of the Swertiinae (Gentianaceae-Gentianeae): Homoplasy and the principle of variable proportions. Syst Bot. 2002;27:548–72.
Xue CY, He TN, Li DZ. Floral nectaries in Swertia: anatomy and morphology. Acta Bot Yunnanica. 2002;24(3):359–69.
Nemomissa S. Floral character states of the Northeast and Tropical East African Swertia species (Gentianaceae). Nord J Bot. 1997;17(2):145–56.
Yuan MY, Küpfer P, Zeltner L. Chromosomal evolution of Gentiana and Jaeschkea (Gentianaceae), with further documentation of chromosome data for 35 species from western China. Plant Syst Evol. 1998;210(3–4):231–47.
Nilsson S. Pollen morphological studies in the Gentianaceae-Gentianinae. Grana Palynologica. 1967;7(1):46–143.
Kamel EA, Elwan ZA, Loutfy MHA. On the taxonomy of Gentiana L. and Gentianella Moench. (Gentianaceae): evidence from seed coat morphology and seed protein electrophoresis. Assiut Univ J Bot. 2006;35(2):1–13.
Kataeva TN, Prokopyev AS, Akinina AA, Chernova OD. Seed morphology of some species in the family Gentianaceae. Biosci Biotechnol Res Asia. 2015;12(3):2287–93.
Whitlock BA, Silver J, Prince JS. Seed coat morphology in Gentianopsis (Gentianaceae). Rhodora. 2010;112(949):58–79.
Clegg MT, Gaut BS, Learn GH Jr, Morton BR. Rates and patterns of chloroplast DNA evolution. Proc Natl Acad Sci USA. 1994;91:6795–801.
Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Muller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, et al. Analysis of genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 2007;104:19369–74.
Moore MJ, Bell CD, Soltis PS, Soltis DE. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc Natl Acad Sci USA. 2007;104:19363–8.
Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci USA. 2010;107:4623–8.
Zhong B, Yonezawa T, Zhong Y, Hasegawa M. The position of Gnetales among seed plants: overcoming pitfalls of chloroplast phylogenomics. Mol Biol Evol. 2010;27:2855–63.
Xi Z, Ruhfel BR, Schaefer H, Amorim AM, Sugumaran M, Wurdack KJ, Endress PK, Matthews ML, Stevens PF, Mathews S, Davis CC. Phylogenomics and a posteriori data partitioning resolve the Cretaceous angiosperm radiation Malpighiales. Proc Natl Acad Sci USA. 2012;109:17519–24.
Barrett CF, Specht CD, Leebens-Mack J, Stevenson DW, Zomlefer WB, Davis JI. Resolving ancient radiations: can complete plastid gene sets elucidate deep relationships among the tropical gingers (Zingiberales)? Ann Bot. 2014;113:119–33.
Ruhfel BR, Gitzendanner MA, Soltis PS, Soltis DE, Burleigh JG. From algae to angiosperms-inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol Biol. 2014;14:23.
Cao Q, Gao QB, Ma XL, Zhang FQ, Xing R, Chi XF, Chen SL. Plastome structure, phylogenomics and evolution of plastid genes in Swertia (Gentianaceae) in the Qing-Tibetan Plateau. BMC Plant Biol. 2022;22:195.
Xu X, Li JP, Chu R, Luan MJ, Wang HY, Song KX, Wei SX, Shi YH, Zhu SX, Wei Z. Comparative and phylogenetic analyses of Swertia L. (Gentianaceae) medicinal plants (from Qinghai, China) based on complete chloroplast genomes. Genet Mol Res. 2022;45:e20210092.
Yang LC, Li JJ, Zhou GY. Comparative chloroplast genome analyses of 23 species in Swertia L. (Gentianaceae) with implications for its phylogeny. Front Genet. 2022;13:895146.
Du Q, Li J, Wang LQ, Chen HM, Jiang M, Chen ZE, Jiang CAB, Gao HD, Wang B, Liu C. Complete chloroplast genomes of two medicinal Swertia species: the comparative evolutionary analysis of Swertia genus in the Gentianaceae family. Planta. 2022;256:73.
Hu Y, Woeste KE, Zhao P. Completion of the chloroplast genomes of five Chinese Juglans and their contribution to chloroplast phylogeny. Front Plant Sci. 2017;7:1955.
Hu H, Al-Shehbaz I, Sun Y, Hao G, Wang Q, Liu J. Species delimitation in Orychophragmus (Brassicaceae) based on chloroplast and nuclear DNA barcodes. Taxon. 2015;64:714–26.
Hu H, Hu QJ, Al-Shehbaz I, Luo X, Zeng TT, Guo XY, Liu JQ. Species Delimitation and Interspecific Relationships of the Genus Orychophragmus (Brassicaceae) Inferred from Whole Chloroplast Genomes. Front Plant Sci. 2016;7:1826.
Zhang YJ, Du LW, Liu A, Chen JJ, Wu L, Hu WM, Zhang W, Kim K, Lee SC, Yang TJ, Wang Y. The complete chloroplast genome sequences of five Epimedium species: lights into phylogenetic and taxonomic analyses. Front Plant Sci. 2016;7:306.
Niu ZT, Xue QY, Zhu SY, Sun J, Liu W, Ding XY. The complete plastome sequences of four orchid species: insights into the evolution of the Orchidaceae and the utility of plastomic mutational hotspots. Front Plant Sci. 2017;8:715.
Wicke S, Schneeweiss GM, Depamphilis CW, Müller KF, Quandt D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 2011;76:273–97.
Tonti-Filippini J, Nevill PG, Dixon K, Small I. What can we do with 1000 plastid genomes? Plant J. 2017;90:808–18.
Zhang L, Wang S, Su C, Harris AJ, Zhao L, Su N, Wang JR, Duan L, Chang ZY. Comparative chloroplast genomics and phylogenetic analysis of Zygophyllum (Zygophyllaceae) of China. Front Plant Sci. 2021;12:723622.
Yang J, Yue M, Niu C, Ma XF, Li ZH. Comparative Analysis of the Complete Chloroplast Genome of Four Endangered Herbals of Notopterygium. Genes. 2017;8:124.
Zhao DN, Ren Y, Zhang JQ. Conservation and innovation: Plastome evolution during rapid radiation of Rhodiola on the Qinghai-Tibetan Plateau. Mol Phylogenet Evol. 2020;144:106713.
Hu GL, Cheng LL, Huang WG, Cao QC, Zhou L, Jia WS, Lan YP. Chloroplast genomes of seven species of Coryloideae (Betulaceae): Structures and comparative analysis. Genome. 2020;63:337–48.
Huang R, Xie X, Li F, Tian EW, Chao Z. Chloroplast genomes of two Mediterranean Bupleurum species and the phylogenetic relationship inferred from combined analysis with East Asian species. Planta. 2021;253:81.
Chen XC, Li QS, Li Y, Qian J, Han JP. Chloroplast genome of Aconitum barbatum var. puberulum (Ranunculaceae) derived from CCS reads using the PacBio RS platform. Front Plant Sci. 2015;6:42.
Dong BR, Zhao ZL, Ni LH, Wu JR, Danzhen ZG. Comparative analysis of complete chloroplast genome sequences within Gentianaceae and significance of identifying species. Chin Tradit Herb Drugs. 2020;51(6):1641–9.
Rogalski M, Schöttler MA, Thiele W, Schulze WX, Bock R. Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions. Plant Cell. 2008;20:2221–37.
Ebert D, Peakall R. Chloroplast simple sequence repeats (cpSSRs): Technical resources and recommendations for expanding cpSSR discovery and applications to a wide array of plant. Mol Ecol Resour. 2009;9:673–90.
George BJ, Bhatt BS, Awasthi M, George B, Singh AK. Comparative analysis of microsatellites in chloroplast genomes of lower and higher plants. Curr Genet. 2015;61:665–77.
Khan G, Zhang FQ, Gao QB, Fu PC, Zhang Y, Chen SL. Spiroides shrubs on Qinghai-Tibetan Plateau: multilocus phylogeography and palaeodistributional reconstruction of Spiraea alpina and S. Mongolica (Rosaceae). Mol Phylogenet Evol. 2018;123:137–48.
Lin M, Qi X, Chen J, Sun L, Zhong Y, Fang J, Hu C. The complete chloroplast genome sequence of Actinidia arguta using the PacBio RS II platform. PLoS ONE. 2018;13:e0197393.
Mehmood F, Abdullah, Ubaid Z, Bao Y, Poczai P. Comparative Plastomics of Ashwagandha (Withania, Solanaceae) and identification of mutational hotspots for barcoding medicinal plants. Plants. 2020;9:752.
Kim SC, Lee JW, Choi BK. Seven complete chloroplast genomes from Symplocos: genome organization and comparative analysis. Forests. 2021;12:608. https://doi.org/10.3390/f12050608.
Ochoterena H. Homology in coding and noncoding DNA sequences: a parsimony perspective. Plant Syst Evol. 2009;282:151–68.
Lopez JL, Lozano MJ, Lagares A, Fabre ML, Draghi WO, Del Papa MF, Pistorio M, Becker A, Wibberg D, Schluter A, Puhler A, Blom J, Goesmann A, Lagares A. Codon usage heterogeneity in the multipartite prokaryote genome: selection-based coding bias associated with gene location, expression level, and ancestry. mBio. 2019;10(3):e00505-19.
Zhao Y, Liu Z, Yang P, Cheng Y, Yang Y. Codon bias analysis method and research progress on codon bias in Camellia sinensis. J Tea Commun. 2016;43:3–7.
Shang M, Liu F, Hua J, Wang K. Analysis on codon usage of chloroplast genome of Gossypium hirsutum. Sci Agric Sin. 2011;44:245–53.
Qin Z, Zheng YJ, Gui LJ, Xie GA, Wu YF. Codon usage bias analysis of chloroplast genome of camphora tree (Cinnamomum camphora). Guihaia. 2018;38(10):1346–55.
Rehman U, Sultana N, Abdullah, Jamal A, Muzaffar M, Poczai P. Comparative chloroplast genomics in Phyllanthaceae species. Diversity. 2021;13(9):403.
Wang Z, Xu B, Li B, Zhou Q, Wang G, Jiang X, Wang C, Xu Z. Comparative analysis of codon usage patterns in chloroplast genomes of six Euphorbiaceae species. PeerJ. 2020;8:e8251.
Zhang Y, Nie X, Jia X, Ding C, Biradar S, Le W, Che X, Song W. Analysis of codon usage patterns of the chloroplast genomes in the Poaceae family. Aust J Bot. 2012;60:461–70.
Zhou T, Wang J, Jia Y, Li WL, Xu FS, Wang XM. Comparative Chloroplast Genome Analyses of Species in Gentiana section Cruciata (Gentianaceae) and the Development of Authentication Markers. Int J Mol Sci. 2018;19:1962.
Chang YJ, Cao YF, Zhang JM, Tian LM, Dong XG, Zhang Y, Qi D, Zhang XS. Study on chloroplast DNA diversity of cultivated and wild pears (Pyrus L.) in Northern China. Tree Genet Genomes. 2017;13(2):44.
Hu GL, Cheng LL, Huang WG, Cao QC, Zhou L, Jia WS, Lan YP. Chloroplast genomes of seven species of Coryloideae (Betulaceae): structures and comparative analysis. Genome. 2020;63(7):337–48.
Dong W, Xu C, Li C, Sun J, Zuo Y, Shi S, Cheng T, Guo J, Zhou S. ycf1, the most promising plastid DNA barcode of land plants. Sci Rep. 2015;5:8348.
Ho TN, James SP. “Gentianaceae”. In: Wu ZY, Raven PH editors, Flora of China. Beijing: Science Press & Missouri: Missouri Botanical Garden Press; 1995. p. 1–139.
Xue CY, Ho TN, Li DZ. Embryology of Swertia (Gentianaceae) relative to taxonomy. Bot J Linn Soc. 2007;155(3):383–400.
Zachos J. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001;292(5517):686–93.
Wolfe JA. A Paleobotanical interpretation of tertiary climates in the Northern Hemisphere: data from fossil plants make it possible to reconstruct Tertiary climatic changes, which may be correlated with changes in the inclination of the earth’s rotational axis. Am Sci. 1978;66(6):694–703.
Miller KG, Fairbanks RG. Evidence for Oligocene-middle Miocene abyssal circulation changes in the western North Atlantic. Nature. 1983;306(5940):250–3.
Keller G, Barron JA. Paleoceanographic implications of Miocene deep-sea hiatuses. GSA Bull. 1983;94(5):590–613.
Buchardt B. Oxygen isotope palaeotemperatures from the tertiary period in the North Sea area. Nature. 1978;275(5676):121–3.
Wang B, Gao L, Su YJ, Wang T. Adaptive evolutionary analysis of chloroplast genes in Euphyllophytes based on complete chloroplast genome sequences. Acta Sci Nat Univ Sunyatseni. 2012;51(3):108–14.
Hartshorne RS, Kern M, Meyer B, Clarke TA, Karas M, Richardson DJ, Simon J. A dedicated haem lyase is required for the maturation of a novel bacterial cytochrome c with unconventional covalent haem binding. Mol Microbiol. 2007;64:1049–60.
Xie Z, Merchant S. The plastid-encoded ccsA gene is required for heme attachment to chloroplast c-type cytochromes. J Biol Chem. 1996;271(9):4632–9. https://doi.org/10.1074/jbc.271.9.4632.
Duarte JM, Wall PK, Edger PP, Landherr LL, Ma H, Pires PK, Leebens-Mack J, Pamphilis CW. Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evol Biol. 2010;10:61.
Soltis DE, Albert VA, Leebens-Mack J, Bell CD, Paterson AH, Zheng CF, Sankoff D, Depamphilis CW, Wall P, Soltis PS. Polyploidy and angiosperm diversification. Am J Bot. 2009;96(1):336–48.
Lemmon AR, Lemmon EM. High-Throughput Identification of Informative Nuclear Loci for Shallow-Scale Phylogenetics and Phylogeography. Syst Biol. 2012;61(5):745–61.
Lemmon EM, Lemmon AR. High-throughput genomic data in systematics and Phylogenetics. Annu Rev Ecol Evol S. 2013;44(1):99–121.
Bogarín D, Pérez-Escobar OA, Groenenberg D, Holland SD, Karremans AP, Lemmon EM, Lemmon AR, Pupulin F, Smets E, Gravendeel B. Anchored hybrid enrichment generated nuclear, plastid and mitochondrial markers resolve the Lepanthes horrida (Orchidaceae: Pleurothallidinae) species complex. Mol Phylogenet Evol. 2018;129:27–47.
Doyle J. DNA protocols for plants-CTAB total DNA isolation. In Hewitt GM, Johnston A, editors. Molecular techniques in taxonomy. Berlin: Springer; 1991.
Patel RK, Jain M. NGS qc toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE. 2012;7:e30619.
Bakker FT, Lei D, Yu JY, Mohammadin S, Wei Z, van de Kerke S, Gravendeel B, Nieuwenhuis M, Staats M, Alquezar-Planas DE. Herbarium genomics: Plastome sequence assembly from a range of herbarium specimens using an iterative organelle genome assembly pipeline. Biol J Linn Soc. 2016;117:33–43.
Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. Using SPAdes de novo assembler. Curr Protoc Bioinforma. 2020;70(1):e102.
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–9.
Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45:W6–11.
Qu XJ, Moore MJ, Li DZ, Yi TS. PGA: A software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 2019;15:50.
Marc L, Oliver D. Sabine K and Ralph B Organellar Genome DRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013;41:575–81.
Kazutaka K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–80.
Dwivedi B, Gadagkar SR. Phylogenetic inference under varying proportions of indel-induced alignment gaps. BMC Evol Biol. 2009;9:211.
Beier S, Thiel T, Münch T, Scholz U, Mascher M. MISA-web: A web server for microsatellite prediction. Bioinformatics. 2017;33:2583–5.
Sun X, Yang Q, Xia X. An improved implementation of effective number of codons (nc). Mol Biol Evol. 2013;30:191–6. https://doi.org/10.1093/molbev/mss201.
Amiryousefi A, Hyvönen J, Poczai P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34(17):3030–1.
Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. Vista: computational tools for comparative genomics. Nucleic Acids Res. 2004;32(Suppl.2):W273–9.
Zhang YJ, Ma PF, Li DZ. High-throughput sequencing of six bamboo chloroplast genomes: phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae). PLoS ONE. 2011;6:e20596.
Lawrie DS, Messer PW, Hershberg R, Petrov DA. Strong purifying selection at synonymous sites in D melanogaster. PLoS Genet. 2013;9:e1003527.
Wang DP, Zhang YB, Zhang Z, Zhu J, Yu J. KaKs_Calculator 2.0: a Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genom Proteom Bioinf. 2010;8(1):77–80.
Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat Methods. 2012;9:772.
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–42.
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian Phylogenetics using tracer 1.7. Syst Biol. 2018;67(5):901–4.
Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–73.
Graham A. Lisianthius pollen from the Eocene of panama. Ann Mo Bot Gard. 1984;71(4):987–93.
Matuszak S, Muellner⁃Riehl AN, Sun H, Favre A. Dispersal routes between biodiversity hotspots in Asia: the case of the mountain genus Tripterospermum (Gentianinae, Gentianaceae) and its close relatives. J Biogeogr. 2016;43(3):580–90.
Rambaut A, Drummond AJ. TreeAnnotator version 1.6.1 [computer program]. http://beast.bio.ed.ac.uk.
Rambaut, A. FigTree v.1.4.4. Available at: http://tree.bio.ed.ac.uk/software/figtree/. (Accessed 25 Oct 2020). 2018.
Acknowledgements
We would like to thank Miss. Jingjing Li and Mr. Hongcai Yue for their help in collection of samples.
Funding
This work was supported by funds from the Qinghai Province Key Laboratory construction project [2022-ZJ-Y18]. The funders were not involved in the study design, data collection, and analysis, decision to publish, or manuscript preparation.
Author information
Authors and Affiliations
Contributions
YL conceived the study, performed data analysis and drafted the manuscript; DS collected samples; ZY and DQ extracted DNA for nextgeneration sequencing; DS, ZY and DQ reviewed the manuscript critically. All authors have read and agreed with the contents of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The authors declared that experimental research works on the plants described in this paper comply with institutional, national and international guidelines. Field studies were conducted in accordance with local legislation and get permissions from provincial department of forest and grass of Qinghai province. Voucher specimens of all plants were deposited at the herbarium of the QTPMB (Qinghai-Tibetan Plateau Museum of biology), Xining, Qinghai Province, China.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Table S1. Genes content of 34 species in 9 genera from Subtribe Swertiinae.
Additional file 2:
Table S2. Length of genes with introns in the cp genomes of 34 species of 9 genus in Subtribe Swertiinae.
Additional file 3:
Table S3. SSR analysis for the cp genome of 34 species in 9 genus of Subtribe Swertiinae.
Additional file 4:
Table S4. Relative synonymous codon usage statistics.
Additional file 5:
Table S5. The GenBankaccession numbers of Subtribe Swertiinae species complete chloroplast genomesthat were downloaded from GenBank
Additional file 6: Figure S1.
Phylogenetic tree of 34Subtribe Swertiinaespecies using Bayesian inference (BI) analysesbased on protein-codinggene sequences. Numbers at nodes correspond to Bayesian inference (BI) posterior probabilities (only probabilities ≥0.95 are shown).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yang, L., Deng, S., Zhu, Y. et al. Comparative chloroplast genomics of 34 species in subtribe Swertiinae (Gentianaceae) with implications for its phylogeny. BMC Plant Biol 23, 164 (2023). https://doi.org/10.1186/s12870-023-04183-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-023-04183-1