Abstract
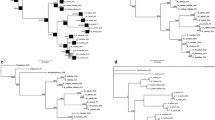
The internal transcribed spacer (ITS) regions of 18S–25S nuclear ribosomal DNA from representatives of 23 species of the subtribeGentianinae and one outgroup species (Centaurium capitatum) were analyzed by polymerase chain reaction amplification and direct DNA sequencing. Within the taxa analyzed, the length of the ITS1 region varied from 221 to 233 bp, ITS2 from 226 to 234 bp. Of the aligned sequences of 497 positions, 151 sites involved gaps or nucleotide ambiguity, 133 were invariable and 213 showed divergence. In pairwise comparisons among the taxa of the subtribeGentianinae and the outgroup, sequence divergence ranged from 1.3% to 34.1% in ITS1, from 0 to 28.1% in ITS2 and from 0.6% to 27.5% in combined ITS1 and ITS2. Phylogenetic trees generated from ITS sequences were highly resolutive and principally concordant with morphological classifications for the major phylogenetic divisions in the subtribe. An ancient divergence leading to two evolutionary lines was suggested in the subtribe by both DNA sequence and morphological data. One line encompasses the generaGentiana, Crawfurdia andTripterospermum, morphologically characterized by their glands on the base of ovary and their plicate corolla, while the other line involves all other members of the subcribe surveyed, characterized by their epipetalous glands and simple corolla without plicae.Megacodon, with glands on the base of ovary but without plicae on its corolla, was revealed to be more related to the latter group than to the former.Comastoma, Gentianella andGentianopsis were shown to be well-defined monophyletic genera.Pterygocalyx showed much closer affinity toGentianopsis than to any other genus. Some conflictions were detected in the genusSwertia.
Similar content being viewed by others
References
Baldwin, B. G., 1992: Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants: an example from theCompositae. — Mol. Phyl. Evol.1: 3–16.
—, 1993: Molecular phylogenetics ofCalycadenia (Compositae) based on ITS sequences of nuclear ribosomal DNA: chromosomal and morphological evolution reexamined. — Amer. J. Bot80: 222–238.
Chase, M. W., Hills, H. H., 1991: Silica gel: an ideal material for field preservation of leaf samples for DNA studies. — Taxon40: 215–220.
Doyle, J. J., 1993: DNA, phylogeny, and the flowering of plant systematics. — Biosci.43: 380–389.
—, 1987: A rapid DNA isolation method for small quantities of fresh tissues. — Phytochem. Bull.19: 11–15.
Gielly, L., 1994: ADN chloroplastique et phylogénies intragénériques. — Ph.D. Thesis, Université Joseph Fourier, Grenoble.
—, 1994: The use of chloroplast DNA to resolve plant phylogenies: noncoding versus rbcL sequences. — Mol. Biol. Evol.11: 769–777.
Gilg, E., 1895:Gentianaceae. — InEngler, A., Prantl, K., (Eds): Die natürlichen Pflanzenfamilien4(2), pp. 50–62. — Leipzig: Engelmann.
Gillett, J. M., 1957: A revision of the North American species ofGentianella Moench. — Ann. Missouri Bot. Gard.44: 195–269.
Hamby, R. K., Zimmer, E. A., 1992: Ribosomal RNA as a phylogenetic tool in plant systematics. — InSoltis, P. S., Soltis, D. E., Doyle, J. J., (Eds): Molecular systematics of plants, pp. 50–91. — New York: Chapman & Hall.
Higgins, D. G., Bleasby, A. J., Fuchs, R., 1992: CLUSTAL:A new multiple sequence alignment program. — Comp. Appl. Biosci.8: 189–191.
Hillis, D., Dixon, M. T., 1991: Ribosomal DNA: molecular evolution and phylogenetic inference. — Quart. Rev. Biol.66: 411–453.
Ho, T. N., 1988:Gentianaceae. — InHo, T. N., (Ed.): Flora Reipublicae Popularis Sinicae62. — Beijing: Science Press.
—, 1990: The infrageneric classification ofGentiana (Gentianaceae). — Bull. British Mus. Nat. Hist., Bot.,20: 169–192.
Holub, J., 1973: New names in Phanerogamae 2. — Folia Geobot. Phytotax.8: 155–179.
Iltis, H. H., 1965: The genusGentianopsis (Gentianaceae): transfer and phytogeographic comments. — Sida2: 129–154.
Kim, K.-J., Jansen, R. K., 1994: Comparisons of phylogenetic hypotheses among different data sets in dwarf dandelions (Krigia, Asteraceae): additional information from internal transcribed spacer sequences of nuclear ribosomal DNA. — Pl. Syst. Evol.190: 157–185.
Löve, A., Löve, D., 1975: The Spanish gentians. — Anal. Inst. Bot. Cavanilles32: 221–232.
- - 1976: The natural genera ofGentianaceae. — InKachroo, P., (Ed.): Recent advances in botany, Prof.P. N. Mehra commemorative volume, pp. 205–222. — Dehra Dun.
Marquand, C. V. B., 1931: New Asiatic gentians: II. — Bull. Misc. Inf. Roy. Bot. Gard. Kew1931: 68–88.
—, 1937: The gentians of China. — Bull. Misc. Int. Roy. Bot. Gard. Kew1937: 134–180.
Pringle, J. S., 1978: Sectional and subgeneric names inGentiana (Gentianaceae). — Sida7: 232–247.
Smith, H., 1965: Notes onGentianaceae. — Notes Roy. Bot. Gard. Edinburgh26(2: 237–258.
—, 1967: InNilsson, S.,: Pollen morphological studies in theGentianaceae. —Gentianinae. — Grana Palynol.7: 46–145.
—, 1970: — InNilsson, S.,: Pollen morphological studies in theGentianaceae. — Acta Univ. Upsaliensis, Abstr. Uppsala Dissert. Sci.165: 5–18.
Suh, Y., Thien, L. B., Reeve, H. E., Zimmer, E. A., 1993: Molecular evolution and phylogenetic implications of internal transcribed spacer sequences of ribosomal DNA inWinteraceae. — Amer. J. Bot.80: 1042–1055.
Swofford, D. L., 1991: PAUP: phylogenetic analysis using parsimony, version 3.0s. — Computer software. — Champaign, Il.: Illinois Natural History Survey.
Sytsma, K. J., Schaal, B., 1985: Phylogenetics of theLisianthus skinneri (Gentianaceae) species complex in Panama utilizing DNA restriction fragment analysis. — Evolution39: 594–608.
—, 1990: Ribosomal DNA variation within and among individuals ofLisianthus (Gentianaceae) populations. — Pl. Syst. Evol.170: 97–106.
Toyokuni, H., 1963: Conspectus Gentianacearum Japonicarum, a general view of theGentianaceae indigenous to Japan. — J. Fac. Sci. Hokkaido Univ., Ser. V,7(4: 137–259.
—, 1965: Systema gentianinarium novissimum — facts and speculation relating to the phylogeny ofGentiana, sensu lato and related genera. — Symb. Asahikawensis1: 147–158.
Tutin, T. G., 1972:Gentiana. — InTutin, T. G. & al., (Eds): Flora Europaea3: 59–63. — Cambridge: Cambridge University Press.
Wojciechowski, M. F., Sanderson, M. J., Baldwin, B. G., Donoghue, M. J., 1993: Monophyly of aneuploidAstragalus (Fabaceae): evidence from nuclear ribosomal DNA internal transcribed spacer sequences. — Amer. J. Bot.80: 711–722.
Yokota, Y., Kawata, T., Iida, Y., Kato, A., Tanifuji, S., 1989: Nucleotide sequences of the 5.8S rRNA gene and internal transcribed spacer regions in carrot and broad bean ribosomal DNA. — J. Mol. Evol.29: 294–301.
Yuan, Y.-M., 1993: Karyological studies onGentiana sect.Cruciata (Gentianaceae) from China. — Caryologia46: 99–114.
—, 1993a: Karyological studies ofGentianopsis Ma and some related genera ofGentianaceae from China. — Cytologia58: 115–123.
—, 1993b: Karyological studies onGentiana sect.Frigida s. l. and sect.Stenogyne (Gentianaceae) from China. — Bull. Soc. Neuchâtel. Sci. Nat.116(2: 65–78.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Yuan, YM., Küpfer, P. Molecular phylogenetics of the subtribeGentianinae (Gentianaceae) inferred from the sequences of internal transcribed spacers (ITS) of nuclear ribosomal DNA. Pl Syst Evol 196, 207–226 (1995). https://doi.org/10.1007/BF00982961
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00982961