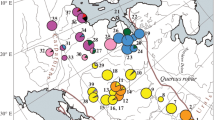
Abstract
Eight pairs of chloroplast DNA (cpDNA) universal primers selected from 34 pairs were used to assess the genetic diversity of 132 pear accessions in Northern China. Among them, six amplified cpDNA fragments showed genetic diversity. A total of 24 variable sites, including 1 singleton variable site and 23 parsimony informative sites, as well as 21 insertion-deletion fragments, were obtained from the combined cpDNA sequences (5309–5535 bp). Two trnL-trnF-487 haplotypes, five trnL-trnF-413 haplotypes, five rbcL haplotypes, six trnS-psbC haplotypes, eight accD-psaI haplotypes and 12 rps16-trnQ haplotypes were identified among the individuals. Twenty-one haplotypes were identified based on the combined fragments. The values of nucleotide diversity (Pi), average number of nucleotide differences (k) and haplotype diversity (Hd) were 0.00070, 3.56408 and 0.7960, respectively. No statistical significance was detected in Tajima’s D test. Remarkably, the important cpDNA haplotypes and their representing accessions were identified clearly in this study. H_19 was considered as one of the ancient haplotypes and was a divergent centre. H_16 was the most common haplotype of the wild accessions. H_2 was the haplotype representing the most pear germplasm resources (46 cultivars and two wild Ussurian Pear accessions), followed by haplotype H_5 (30 cultivars, two wild Ussurian Pear accessions and four sand pears in outgroups) representing the cultivars ‘Dangshan Suli’ and ‘Yali’, which harbour the largest and the second largest cultivation areas in China. More importantly, this study demonstrated, for the first time, the supposed evolution routes of Pyrus based on cpDNA divergence in the background of pear phylogeny in Northern China.






Similar content being viewed by others
References
Bao L, Chen K, Zhang D, Cao Y, Yamamoto T, Teng Y (2007) Genetic diversity and similarity of pear cultivars native to East Asia revealed by SSR (simple sequence repeat) markers. Genet Resour Crop Evol 54:959–971
Bao L, Chen K, Zhang D, Li X, Teng Y (2008) An assessment of genetic variability and relationships within Asian pears based on AFLP (amplified fragment length polymorphism) markers. Sci Hortic 116:374–380
Bell R, Quamme H, Layne R, Skirvin R (1996) Pears. In: Janick J, Moore JN (eds) Fruit breeding, vol 1: tree and tropical fruits. Wiley, New York, pp 441–514
Cao Y (2014) Pear varieties in China. China Agriculture Press, Beijing (in Chinese)
Cao Y, Liu F, Gao Y, Jiang L, Wang K, Ma Z, Zhang K (2007) SSR analysis of genetic diversity of pear cultivars. Acta Horticulturae Sinica 34:305–310 (in Chinese)
Cao Y, Tian L, Gao Y, Liu F (2012) Genetic diversity of cultivated and wild Ussurian Pear (Pyrus ussuriensis Maxim.) in China evaluated with M13-tailed SSR markers. Genet Resour Crop Evol 59:9–17
Challice J, Westwood M (1973) Numerical taxonomic studies of the genus Pyrus using both chemical and botanical characters. Bot J Linn Soc 67(2):121–148
Chang Y, Cao Y, Zhang J, Tian L, Dong X, Zhang Y, Qi D (2014) Studies on genetic diversity of pear germplasm resources in Liaoning province of China based on chloroplast DNA analysis. Acta Horticulturae Sinica 41(7):1307–1316 (in Chinese)
Clegg M, Zurawski G (1992) Chloroplast DNA and the study of plant phylogeny: present status and future prospects. In: Soltis PS, Soltis DE, Doyle JJ (eds) Molecular systematics of plants. Chapman and Hall, New York, pp 1–13
Clement M, Posada D, Crandall K (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Cornuet J, Pudlo P, Veyssier J, Dehne-Garcia A, Gautier M, Leblois R, Marin J, Estoup A (2014) DIYABC v2.0: a software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 30(8):1187–1189
Demesure B, Sodzi N, Petit R (1995) A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Mol Ecol 4:129–134
Downie S, Palmer J (1992) Use of chloroplast DNA rearrangements in reconstructing plant phylogeny. In: Soltis PS, Soltis DE, Doyle JJ (eds) Molecular systematics of plants. Chapman and Hall, New York, pp 14–35
Doyle J, Doyle J (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19:11–15
Doyle J, Davis J, Soreng R, Garvin D, Anderson M (1992) Chloroplast inversion and the origin of the grass family (Poaceae). Proc Natl Acad Sci U S A 89:7722–7726
Dumolin-Lapegue S, Pemonge M, Petit R (1997) An enlarged set of consensus primers for the study of organelle DNA in plants. Mol Ecol 6:393–397
He Z, Wu B, Yang Q (2011) Identification of Pyrus sinkiangensis and classification discussion of ‘Korla pear’. Shanxi Fruits 5:36–39 (in Chinese)
Iketani H, Manabe T, Matsuta N, Akihama T, Hayashi Y (1998) Incongruence between RFLPs of chloroplast DNA and morphological classification in east Asian pear (Pyrus spp). Genet Resour Crop Evol 45:533–539
Jing Z (1989) The origin and development of Pingguoli. Northern Horticulture 1:21–22 (in Chinese)
Katayama H, Ogihara Y (1996) Phylogenetic affinities of the grasses to other monocots as revealed by molecular analysis of chloroplast DNA. Curr Genet 20:527–581
Katayama H, Uematsu C (2003) Comparative analysis of chloroplast DNA in Pyrus species: physical map and gene localization. Theor Appl Genet 106:303–310
Katayama H, Adachi S, Yamamoto T, Uematsu C (2007) A wide range of genetic diversity in pear (Pyrus ussuriensis var. aromatica) genetic resources from Iwate, Japan revealed by SSR and chloroplast DNA markers. Genet Resour Crop Evol 54:1573–1585
Katayama H, Tachibana M, Iketani H, Zhang S, Uematsu C (2012) Phylogenetic utility of structural alterations found in the chloroplast genome of pear: hypervariable regions in a highly conserved genome. Tree Genet Genomes 8:313–326
Kelchner S, Clark L (1997) Molecular evolution and phylogenetic utility of the chloroplast rpII6 intron in Chusquea and the Bambusoideae (Poaceae). Mol Phylogenet Evol 8:385–397
Kimura T, Iketani H, Kotobuki K, Matsuta N, Ban Y, Hayashi T, Yamamoto T (2003) Genetic characterization of pear varieties revealed by chloroplast DNA sequences. J Hortic Sci Biotechnol 78:241–247
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Liu Y (2006) Research on chloroplast DNA diversity of genus pear in China. Master Dissertation. Capital Normal University, Beijing. (in Chinese)
Liu J, Zheng X, Potter D, Hu C, Teng Y (2012) Genetic diversity and population structure of Pyrus calleryana (Rosaceae) in Zhejiang Province, China. Biochem Syst Ecol 45:69–78
Liu J, Sun P, Zheng X, Potter D, Li K, Hu C, Teng Y (2013) Genetic structure and phylogeography of Pyrus pashia L. (Rosaceae) in Yunnan Province, China, revealed by chloroplast DNA analyses. Tree Genet Genomes 9(2):433–441
Lu F, Zhang Y (2009) Taxonomic status identification of Ping guo pear by AFLP molecular marker. J Anhui Agric Sci 37:1937–1938 (in Chinese)
Lu J, Wu J, Zhang S, Wu H, Zhang Y (2011) Genetic diversity and polygentic relationship among pears revealed by SSR analysis. J Nanjing Agric Univ 34(2):38–46 (in Chinese)
Ma Y, Zhang Y (2009) The application of RAPD in status identification of ‘Pingguoli’. Chin Agric Sci Bull 25:71–73 (in Chinese)
Ma B, Niu J, Pan L, Feng J, Lu X (2004a) Study on the genetic relationships among 18 pear cultivars by RAPD. J Fruit Sci 21(6):521–525 (in Chinese)
Ma B, Niu J, Wu Z, Tan W (2004b) Molecular markers analysis of main cultivars relationship between Pyrus in Xinjiang. J Shihezi Univ (Nat Sci) 22(2):97–102 (in Chinese)
Montanari S, Saeed M, Knäbel M, Kim Y, Troggio M, Malnoy M et al (2013) Identification of Pyrus single nucleotide polymorphisms (SNPs) and evaluation for genetic mapping in European pear and interspecific Pyrus hybrids. PLoS One 8(10):e77022
Nei M, Li W (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A 76:5269–5273
Palmer J, Jorgensen R, Thompson W (1985) Chloroplast DNA variation and evolution in Pisum; patterns of change and phylogenetic analysis. Genetics 109:195–213
Parani M, Lakshmi M, Ziegenhagen B, Fladung M, Senthilkumar P, Parida A (2000) Molecular phylogeny of mangroves VII PCR-RFLP of trnS-psbC and rbcL gene regions in 24 mangrove and mangrove-associate species. Theor Appl Genet 100:454–460
Parducci L, Szmidt A (1999) PCR-RFLP analysis of cpDNA in the genus Abies. Theor Appl Genet 98:802–808
Petit R, Kremar A, Wagner D (1993) Geographic structure of chloroplast DNA polymorphisms in European oaks. Theor Appl Genet 87:122–128
Pu F, Wang Y (1963) Pomology of China. Pears, vol 3. Shanghai Science and Technology Press, Shanghai (in Chinese)
Pu F, Lin S, Song W, Chen R, Li X (1985) Research on the karyotypes of Pyrus in China I. Plant Sci J 3(4):381–387 (in Chinese)
Pu F, Lin S, Chen R, Song W, Li X (1986) Research on the karyotypes of Pyrus in China II. Acta Horticulturae Sinica 13(2):87–90 (in Chinese)
Qu Z, Pan J, Shan Z (1990) The log of Beijing fruit tree. Beijing Press, Beijing, pp 239–240 (in Chinese)
Qu B, Jin X, Chen Y, Liu H, Wang P (2001) RAPD analysis of germplasm resources in Pyrus. Acta Horticulturae Sinica 28:460–462 (in Chinese)
Qu B, Jin X, Chen Y, Liu H, Wang P (2002) Classification study of Pingguoli in Yanbian area using RAPD markers. Progress Hortic 5:178–182 (in Chinese)
Qu B, Jin X, Chen Y, Liu H, Wang P, Zheng D (2003) Classification study of Pingguoli in Yanbian area by using RAPD markers. J Jilin Agric Univ 25:292–295 (in Chinese)
Rubtsov G (1944) Geographical distribution of the genus Pyrus and trends and factors in its evolution. Am Nat 78:358–366
Sehic J, Garkava-Gustavsson L, Fernández-Fernández F, Nybom H (2012) Genetic diversity in a collection of European pear (Pyrus communis) cultivars determined with SSR markers chosen by ECPGR. Sci Hortic 145:39–45
Shan J, Li J, Luo S, Cheng Q, Li C (2010) Analysis of genetic relationships of pear germplasm resources in Xinjiang based on ISSR and RAPD. Xinjiang Agric Sci 47(9):1714–1721 (in Chinese)
Shen Y, Teng Y, Tanabe K (2006) RAPD analysis for genetic assessment of some cultivars of Pyrus pyrifolia derived from China and Japan. Acta Horticulturae Sinica 33(3):621–624 (in Chinese)
Small R, Ryburn J, Cronn R, Seelanan T, Wendel J (1998) The tortoise and the hare: choosing between non-coding plastome and nuclear ADH sequences for phylogeny reconstruction in recently diverged plant group. Am J Bot 85:1301–1345
Swofford D (2002) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland MA. (Program)
Taberlet P, Gielly L, Pauton G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17:1105–1109
Tajima F (1989) Statistical method for testing the neutral mutation hypotheses by DNA polymorphism. Genetics 123:585–595
Teng Y, Tanabe K, Tamura F, Itai A (2001) Genetic relationships of pear cultivars in Xinjiang, China as measured by RAPD markers. J Hortic Sci Biotechnol 76:771–779
Teng Y, Tanabe K, Tamura F, Itai A (2002) Genetic relationships of Pyrus species and cultivars native to East Asia revealed by randomly amplified polymorphic DNA markers. J Am Soc Hortic Sci 127:262–270
Urrestarazu J, Royo J, Santesteban L, Miranda C (2015) Evaluating the influence of the microsatellite markerset on the genetic structure inferred in Pyrus communis L. PLoS One 10(9):e0138417
Vavilov N (1951) The origin, variation, immunity and breeding of cultivated plants. Chronica Botanica 13:1–366
Wang Y (1988) Deciduous fruit tree breeding. Press of Agriculture, Beijing, pp 192–193 (in Chinese)
Wu G (1984) The taxonomy of temperate-zone fruit trees in China. Press of Agriculture, Beijing, pp 33–80 (in Chinese)
Wuyun T, Ma T, Uematsu C, Katayama H (2013) A phylogenetic network of wild Ussurian pears (Pyrus ussuriensis Maxim.) in China revealed by hypervariable regions of chloroplast DNA. Tree Genet Genomes 9:167–177
Yamamoto T, Kimura T, Sawamura Y, Kotobuki K, Ban Y, Hayashi T, Matsuta N (2001) SSRs isolated from apple can identify polymorphism and genetic diversity in pear. Theor Appl Genet 102:865–887
Yamamoto T, Kimura T, Sawamura Y, Manabe T, Kotobuki K, Hayashi T, Ban Y, Matsuta N (2002a) Simple sequence repeats for genetic analysis in pear. Euphytica 124:129–137
Yamamoto T, Kimura T, Sawamura Y, Manabe T, Kotobuki K, Hayashi T, Ban Y, Matsuta N (2002b) Genetic linkage maps constructed by using an interspecific cross between Japanese and European pears. Theor Appl Genet 106:9–18
Yang H (1985) The application of palynology in the taxonomy of some plants in Pyrus. Fruit Sci 3:2–9 (in Chinese)
Yang C (2010) Genetic diversity of cytoplasm heredity in some main cultivars of Pyrus. Master Dissertation. Shihezi University, Xinjiang. (in Chinese)
Yang L, Li W, Qu B (2010) The classification discussion of ‘Pingguoli’. J Agric Sci Yanbian Univ 32:73–76 (in Chinese)
Yao L, Zheng X, Cai D, Gao Y, Wang K, Cao Y, Teng Y (2010) Exploitation of Malus EST-NSSRs and the utility in evaluation of genetic diversity in Malus and Pyrus. Genet Resour Crop Evol 57:841–851
Yu D (1979) Taxonomy of the fruit tree in China. Agriculture Press, Beijing (in Chinese)
Zhang J, Chen Q, Cao Y, Yang X, Fan J, Hu H (2016) Genetic diversty and phylogenetics of pear (Pyrus L.) germplasm resources from Hubei province revealed by chloroplast DNA variation. J Genet Resour 17(4):766–772 (in Chinese)
Zong Y, Sun P, Liu J, Yue X, Niu Q, Teng Y (2014) Chloroplast DNA-based genetic diversity and phylogeography of Pyrus betulaefolia (Rosaceae) in Northern China. Tree Genet Genomes 10:739–749
Zou L, Zhang X, Zhang Z, Song B, Guo S (1986) Research on genetic relationships of Pyrus based on pollen morphology. Acta Horticulturae Sinica 13(4):219–224 (in Chinese)
Acknowledgements
The study was sponsored by the National Natural Science Foundation of China (grant No. 31272128).
Data archiving statement
The authors declare that all the work described in this manuscript followed the standard Tree Genetics and Genomes policy. The sequences will be uploaded soon, and all the sequences in this study will be found in the National Center of Biotechnology Information (NCBI) database.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D. Chagné
Rights and permissions
About this article
Cite this article
Chang, YJ., Cao, YF., Zhang, JM. et al. Study on chloroplast DNA diversity of cultivated and wild pears (Pyrus L.) in Northern China. Tree Genetics & Genomes 13, 44 (2017). https://doi.org/10.1007/s11295-017-1126-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-017-1126-z