Abstract
Background
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a central enzyme in glycolysi, we performed genome-wide identification of GAPDH genes in wheat and analyzed their structural characteristics and expression patterns under abiotic stress in wheat.
Results
A total of 22 GAPDH genes were identified in wheat cv. Chinese spring; the phylogenetic and structure analysis showed that these GAPDH genes could be divided into four distinct subfamilies. The expression profiles of GAPDH genes showed tissue specificity all over plant development stages. The qRT-PCR results revealed that wheat GAPDHs were involved in several abiotic stress response.
Conclusions
Wheat carried 22 GAPDH genes, representing four types of plant GAPDHs (gapA/B, gapC, gapCp and gapN). Whole genome duplication and segmental duplication might account for the expansion of wheat GAPDHs. Expression analysis implied that GAPDHs play roles in plants abiotic stress tolerance.
Similar content being viewed by others
Background
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), catalyzing the conversion of glyceraldehyde-3-phosphate (G3P) to 1,3-biphosphoglycerate in the presence of NAD+ and inorganic phosphate, is a key enzyme in the glycolytic pathway [1]. GAPDH was once considered as a simple “housekeeping gene” and has been used as a reference in gene expression and protein studies [2]. However, it’s recently demonstrated that GAPDH plays crucial roles in many cellular processes in addition to glycolysis [3–5].
As a ubiquitous enzyme, GAPDH exists in nearly all organisms. In plant cells, GAPDHs, according to their different subcellular locations, could be classified into three groups as follows: GAPA/B, encoded by gapA and gapB, chloroplastic phosphorylating NADP-specific GAPDHs participating in photosynthetic CO2 fixation; GAPC, from gene gapC, a phosphorylating NAD-dependent GAPDH catalyzing the conversion of glyceraldehyde-3-P (Ga3P) to 1,3-bisphosphoglycerate in cytoplasm; GAPCp, encoded by gapCp, involved in glycolytic energy production in non-green plastids [6–14]. Depending on species, each type of gap gene may be further duplicated, for example, Arabidopsis contains two gapA, two gapC and two gapCp genes, but only a single gapB [15, 16]. In addition, plant contains a cytosolic non-phosphorylating NADP-dependent GAPDH (GAPN), which belongs to the aldehyde dehydrogenase superfamily and has no close functional and structural relationships with phosphorylating GAPDHs, catalyzing the oxidation of Ga3P to 3-phosphoglycerate (3PGA) [17–21].
GAPDH is essentially a tetramer of identical or similar subunits. The glycolytic GAPDHs (GAPC, GAPCp) and A4-GAPDH of oxygenic phototrophs compose of identical subunits, while the main photosynthetic isoform of land plants (A2B2-GAPDH) consist of similar subunits. The main features of GAPDH protein monomers are highly conserved in all living organisms, consisting of an N-terminal NAD (P)-binding domain called Gp_dh_N domain (PF00044) and a C-terminal catalytic domain named Gp_dh_C domain (PF02800). Some GAPDH genes (AtGAPB) contain an extra incomplete CP12 domain (PF02672). While GAPNs contain an Adledh domain.
Besides its pivotal role in glycolysis, GAPDH is a moonlighting protein [13]. In Mammals, GAPDH participatosis [1, 4, 5, 22] and cell toxicity [23]. Nonglycolytic functiones in signal transduction cascades, DNA repair, transcriptional regulation, apopts of plant GAPDH have been demonstrated as well, especially in abiotic stress responses. For instance, In Aspen (Populus tremula), GAPDH showed some increase in response to water deficit [24]. Overexpression of a cytosolic OsGAPC3 enhanced salt tolerance in rice [25]. Furthermore, GAPDH generated immunocomplex with NtOSAK (osmotic stress-activated protein kinase) in salt-treated tobacco cells [26]. Moreover, GAPDH interacted with Phospholipase Dδ (PLDδ) to transduce hydrogen peroxide signals and the GAPC–PLDδ interaction mediated response to ABA and water deficits in Arabidopsis [27]. Besides abiotic stress responses, plant GAPDHs have been implicated in embryo development, pollen development, root development [16, 28–31].
Although vast studies have been conducted on biochemical properties and physiological functions of GAPDHs, few systematic research on the evolution and functional divergence of GAPDH gene family have been conducted, especially in wheat (Triticum aestivum). Recent plant genome sequencing projects have strongly promoted the identification and characterization of plant genes. Wheat genome sequencing is now nearing completion (http://www.wheatgenome.org/). Hence, a comprehensive analysis of the wheat GAPDH family was conducted here. We systematically identified and characterized the GPADH genes of wheat (TaGAPDH) and compared them with GAPDH in Arabidopsis thaliana, Hordeum vulgare, Aegilops tauschii and Triticum urartu. In addition, the expression profiles of GAPDH genes in different tissues and at different stages as well in response to several stress were analyzed. Finally, the inducible expression of wheat GAPDH genes were detected by qRT-PCR experiments, and some GAPDHs were found notably responding to abiotic stress.
Results
Identification, phylogenetic analysis and classification of wheat GAPDH genes
By Hidden Markov model (HMM) searches on the available Triticum aestivum sequenced genome, 40 sequences were identified. 15 of these sequences were removed for absence of complete Gp_dh_N or Gp_dh_C domain and 13 of them harbour both domains (Additional file 1). Then, the 25 putative GAPDHs were used as queries to BLASTn against the contigs and singletons assembled from EST sequences (Additional file 2). Six of them had no matched contig or singleton, suggesting that they were not expressed in wheat. Finally, 19 GAPDHs were identified in wheat (Table 1). These GAPDHs genes located on chromosomes 2, 4, 6 and 7. Chromosomes 2A, 2B, 4B and 4D contained one GAPDH, respectively. Chromosomes 2D, 6B, 6D, 7A, 7B and 7D contained two GAPDHs, respectively. And chromosomes 6A contained 3 GAPDHs (Table 1). Meanwhile, 3 GAPNs that located on chromosomes 2A, 2B and 2D were identified (Table 1 and Additional file 3).
The coding sequences (CDS) and protein sequences of wheat GAPDHs that contain both domains (TaGAPDH1-13) and GAPNs (TaGAPN1-3) were aligned by DNAMAN with pairwise method (Additional files 4 and 5). It showed that the 13 wheat GAPDH genes shared 47.04–99.7 % identity at both CDS and putative amino acid levels. For instance, TaGAPDH1/2, TaGAPDH3/4/6/8, TaGAPDH5/7, TaGAPDH9/11/13 and TaGAPDH10/12 showed high identity (≥90 %) at both CDS and protein sequence levels, respectively. Meanwhile, the 3 GAPNs showed low identity (<30 %) with GAPDHs, but high identity (>90 %) with each other (Additional file 5). Moreover, the molecular weights (MW) and isoelectric points (pI) of wheat GAPDH and GAPN proteins were calculated by ProtParam tool in ExPASy. The results revealed that the MWs of GAPDHs varied from 14.74 to 43.86 kDa, and that of GAPNs varied from 46.65 to 53.07 kDa, the pIs of GAPDHs ranged from 6.40 to 8.65 while that of GAPNs floated around 6.40 (Table 1).
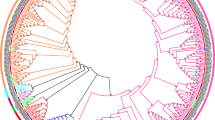
For phylogenetic analysis, classification and nomenclature, Arabidopsis thaliana, Hordeum vulgare, Aegilops tauschii, Triticum urartu and Triticum turgidum GAPDHs and GAPNs gene were identified as well (Additional file 6). And a rooted Neighbor-joining phylogenetic tree was constructed by the MEGA5.1 program with the default parameters (Fig. 1). Combing with the proposed nomenclature and subcellular location of Arabidopsis thaliana GAPDHs, the employed GAPDH genes could be distributed into four subfamilies (Fig. 1). Subfamily I was corresponding to gapA/B gene, subfamily II to gapCp genes, subfamily III to gapC gene, and subfamily IV to gapN gene. In wheat, TaGAPDH1 and TaGAPDH2 were distributed into subfamily I, TaGAPDH5/7/9/11/13 were grouped into subfamily II, TaGAPDH4/6/8/10/12 were clustered in subfamily III, and all TaGAPNs were in subfamily IV (Fig. 1; Additional file 6).
Molecular phylogenetic analysis of GAPDH gene families in Arabidopsis thaliana, Triticum aestivum, Aegilops tauschii, Triticum urartu and Triticum turgidum. The rooted Neighbor-joining phylogenetic tree of GAPDH genes was inferred from the amino acid sequence alignments of GAPDHs by MEGA5.1 with Neighbor-joining method under default parameters. The values on the tree represent bootstrap confidence values inferred from 1000 replicates
Structure and conserved residues analysis of GAPDH
Owing to the transcript annotation of wheat genome, the analysis and comparison of the structural features of GAPDHs in different families were conducted (Fig. 2). It indicated that the organization (number, order and length) of exons were almost conserved within each GAPDH subfamily, while the introns and UTRs showed variable lengths and distribution (Figs. 1 and 2). The majority of GAPDH genes in subfamilies II, III and IV contained 13, 10 and 8 introns, respectively. And in subfamily I, GAPAs and GAPBs mostly contained 8 and 4 introns, respectively. In wheat, GAPDHs in same subfamily shared similar exon/intron structure, for example, TaGAPDH1/2, TaGAPDH4/6/810/12, TaGAPDH5/7/9/11/13 and TaGAPNs shared similar exon/intron structures, respectively (Figs. 1 and 2).
Structure analysis of GAPDH gene families in Arabidopsis thaliana, Triticum aestivum, Aegilops tauschii and Triticum urartu. Gene structures of GAPDHs analyzed by GSDS (http://gsds.cbi.pku.edu.cn/). Exons are shown as green boxes, introns are shown as thin lines, and UTRs are shown as blank boxes
To investigate the conserved residues of GAPDH proteins, we aligned wheat GAPDH protein sequences and detected the conserved sites of them (Additional file 7). The alignment displayed that amino acids structure of TaGAPDHs are fairly conserved and there are several identical regions such as “INGFGRIGR”, “KYD”, “GVF”, “GAKKV” “SNASCTTNCLAP”, “STGAAKAV”, “RVPT”, “VS”, “DF” and “WYDNEWGYS”. Among these regions, the “INGFGRIGR”, “SNASCTTNCLAP”, “RVPT”, “VS”, “DF”, and “WYDNEWGYS” match well with that of animal/fungi GAPDHs alignment (Additional file 8). It suggested that GAPDHs are much conserved during evolution. Furthermore, the alignment of GAPNs showed that GAPNs are nearly identical in different plants (Additional file 9). To give a visual insight to the GAPDH proteins similarity, a motif figure was constructed by submitting amino acid sequences of GAPDH genes to the MEME website (Fig. 3 and Additional file 10). Almost every GAPDH possessed the first six motifs while every GAPNs possessed the last six ones, the motif structure of GAPDH proteins from different species shared high similarity within each subfamily (Figs. 1 and 3). Furthermore, the conserved regions like “SNASCTTNCLAP” and “WYDNEWGYS” were exhibited completely in those motifs, meeting the results of alignment.
Structure of GAPDH proteins in Arabidopsis thaliana, Triticum aestivum, Aegilops tauschii, Triticum urartu and Triticum turgidum. The line represents the coding sequence and 12 motifs are shown. The amino acid sequences of GAPDH were used as an input of MEME. The total number of input GAPDH and GAPN genes is 42 and 9 respectively
Expression analysis of GAPDHs by microarray
To analyze the expression profiles of GAPDHs, the microarray data of Arabidopsis thaliana, Hordeum vulgare GAPDHs were downloaded from the BAR website (http://bar.utoronto.ca/) with their accession numbers or corresponding probe set IDs, the expression data of wheat GAPDHs were downloaded from PLEXdb (http://www.plexdb.org/) (Additional file 6). Combined with the analysis on Genevestigator (https://genevestigator.com/gv/), the tissue expression profiles and the inducible expression profiles in response to stress of GAPDHs were generated (Fig. 4).
Expression profiles of GAPDHs in different tissues. a Expression profiles of Arabidopsis thaliana GAPDHs in different tissues. b Expression profiles of barley GAPDHs in different tissues. c Expression profiles of wheat GAPDHs in different tissues. d Expression profiles of wheat GAPDHs in different tissues at different development stages. Number 1–9 mean germination, seedling growth, tillering, stem elongation, booting, inflorescence emergence, anthesis, milk development, dough development and ripening stages, respectively
GAPDH genes were expressed almost all over the plant developmental stages and showed notable tissue specificity (Fig. 4). In Arabidopsis, AtGAPA1, AtGAPA2 and AtGAPB were significantly highly expressed in leaf but their expression in root were negligible relative to that in leaf. AtGAPCs and AtGAPN were highly expressed in root, leaf and flower. While AtGAPCps showed low expression level in all tissues except relatively high level in root (Fig. 4a). In barley, HvGAPDH1, HvGAPDH3 and HvGAPN shared the same expression pattern that significantly high in leaf but extremely low in other tissues. Meanwhile, HvGAPDH2 slightly and nearly constantly expressed in all tissues, just as AtGAPCps did (Fig. 4a and b). Then in wheat, TaGAPDH1 and TaGAPN1 were strongly expressed in leaf, shoot, crown and inflorescence but the case came out opposite in root. In addition, TaGAPDH4, TaGAPDH7, TaGAPDH9, TaGAPDH10 and TaGAPDH12 were expressed at roughly steady but various levels in all tissues, and among them, TaGAPDH4/12 and TaGAPDH7/9 shared synchronous expression patterns, respectively (Fig. 4c and d).
Expression profiles of GAPDH genes response abiotic stresses
The expression patterns of wheat GAPDH genes under cold, heat, drought and salt treatments were detected by qRT-PCR (Fig. 5b) and that of Arabidopsis were generated with the microarray data from Arabidopsis eFP Browser (Fig. 5a). Their expression profiles appeared to be complex in view of the results.
Expression analysis of GAPDH genes under abiotic stresses. a Expression heatmap of GAPDHs under abiotic stresses in Arabidopsis. The microarray data was downloaded from Arabidopsis eFP Browser. Heatmap was plot by Heatmapper. b Expression heatmap of GAPDHs under abiotic stresses in wheat. The data came from qRT-PCR analysis with 2–ΔΔCt method. Heatmap were generated with MeV v4.9. The relative expression levels were intuitively reflected in the heatmap with the gradient color green/black/red (low to high)
Under cold treatments, in Arabidopsis, AtGAPAs showed slightly increased expression at early stress stage in shoots but were strongly up-regulated in roots. AtGAPB were intensely transcribed in both shoots and roots. Roughly, AtGAPCs showed up-regulated expression in shoots and down-regulated expression in roots. AtGAPCps shared same expression pattern: significantly up-regulated in shoots and down-regulated in roots (Fig. 5a). While in wheat, TaGAPDH1, TaGAPDH6, TaGAPDH9, TaGAPDH10 and TaGAPDH12 were dramatically up-regulated in shoots (Fig. 5b). In roots, TaGAPDH10 and TaGAPDH12 were up-regulated, and TaGAPDH4 and TaGAPDH9 were notably down-regulated (Fig. 5b).
Exposed to heat stress, transcription of most Arabidopsis GAPDHs were reduced in both shoots and roots except for AtGAPB and AtGAPC1 were slightly up regulated in shoots (Fig. 5a). TaGAPDH1, TaGAPDH9, TaGAPDH10 and TaGAPDH12 were up-regulated, and TaGAPDH4 and TaGAPDH6 were down-regulated in shoots. In roots, most of TaGAPDHs were down-regulated except TaGAPDH10 and TaGAPDH12 (Fig. 5b).
During drought treatment, AtGAPAs showed down-regulated expression in shoots and fluctuant expression in roots. AtGAPB were up-regulated at first and down-regulated later in shoots. In roots, AtGAPB were intermittently up and down-regulated. AtGAPCps were prominently up-regulated in shoots and transcribed without sharp variation in roots (Fig. 5a). In wheat shoots, TaGAPDH1, TaGAPDH9 and TaGAPDH12 were found up-regulated. Moreover, TaGAPDH4, TaGAPDH6 and TaGAPDH10 were down regulated. While in roots, at early stage of stress, all these TaGAPDHs except TaGAPDH6 were up-regulated at the transcriptional levels (Fig. 5b).
Immersed in thick salt solution, AtGAPCps in shoots and AtGAPAs and AtGAPB in roots were up regulated as the processing time increasing. Besides, the rest were down regulated (Fig. 5a). In addition, TaGAPDH12 in wheat shoots showed high transcription levels (Fig. 5b). Furthermore, Arabidopsis GAPDHs expression profiles under osmotic stress were similar to that under salt treatment (Fig. 5a).
Discussion
Expansion of GAPDH family in wheat
Polyploidy genomes is a common phenomenon in the plant evolutionary process. Along with the genome polyploidy, the loss and insertion of large segments and the chromosomal rearrangements have occurred frequently on chromosomes, these changes might result in gene expression, silence and loss [32, 33]. Common wheat (Triticum aestivum L; 2n = 6x = 42; AABBDD) is allohexaploid. During its evolutionary process, two polyploidization issues had occurred. First of all, Triticum monococcum (2n = 2x = 14; AA) and Aegilops speltoides (2n = 2x = 14; SS) have hybridized and undergone a natural polyploidization and then been naturalized into Triticum turgidum (4n = 4x = 28; AABB). After that, Triticum turgidum have hybridized with Aegilops tauschii (2n = 2x = 14; DD) and undergone polyploidization once again resulting in the common wheat (2n = 6x = 42; AABBDD) [34, 35]. Theoretically, every wheat genes would have three homoeologous gene, but as a consequence of the side effect of polyploidy, evolutionary and acclimation process, many wheat genes have only one or two homoeologous genes that expressed. Here, 22 GAPDH genes representing four major types of plant GAPDH (gapA/B, gapC, gapCp and gapN) were identified in hexaploid wheat cv. Chinese spring, confirming the demonstration above. The high identity of wheat GAPDHs within and between subfamilies indicated that some highly identical GAPDHs may be generated via duplications (Fig. 2 and Additional file 7). Given that there were 6 and 5 GAPDH genes identified in Aegilops tauschii and Triticum urartu (2n = 2x = 14; AA), respectively (Table 1 and Additional file 6), we inferred that whole genome duplication and segmental duplication have contributed to expansion of GAPDHs family in wheat.
GAPDHs play compelling roles in plant abiotic stress tolerance
Inducible expression analysis here revealed that those investigated GAPDH gene expression could be induced by at least one abiotic stress treatment (cold, heat, drought, osmotic or salt). For example, TaGAPDH12 was found up-regulated significantly in shoots under cold, heat, drought and salt stresses (Fig. 5). It implied that GAPDH may have multiple functions except for metabolism roles in plant, such as involving in abiotic stress resistance. This novel function of GAPDH was ubiquitous in plants. For instance, maize GAPC3 and GAPC4 were induced by anaerobic stress [36], and overexpression of a rice cytosolic gene OsGAPC3 enhances salt tolerance [25]. In Arabidopsis thaliana, GAPCs, transduced the H2O2 signal by interacting with the plasma membrane-associated phospholipase D (PLDδ) and the knockout of GAPCs made plants less responsive to water deficits than the wild type [27].
Taken together, it was demonstrated that GAPDH is a multifunction protein besides its key role in glycolysis, especially in plant abiotic stress resistance. Further study to detect its moonlight function and illuminate concrete mechanism will enhance the understanding of this common but amazing protein.
Conclusions
Wheat carried 22 GAPDH genes, representing four types of plant GAPDHs (gapA/B, gapC, gapCp and gapN). Whole genome duplication and segmental duplication might account for the expansion of wheat GAPDHs. The 22 GAPDHs were distributed on chromosomes 2, 4, 6 and 7. According to phylogenetic analysis and structural characteristics, GAPDHs could be classified into four subfamilies. Microarray analysis showed that GAPDH genes were expressed almost all over the plant developmental stages with notable tissue specificity and involved in several abiotic stresses responses. A further qRT-PCR analysis of wheat GAPDHs indicated that these GAPDH genes followed different expression patterns in response to abiotic stresses. It was speculated that GAPDHs play roles in plants abiotic stress tolerance.
Methods
Database mining and identification of GAPDH genes
Wheat, barley, Aegilops tauschii, and Triticum urartu genome sequences and protein sequences were downloaded from Ensembl Plants (http://plants.ensembl.org/index.html) [34, 37–39]. A local protein database were established with these protein sequences. To investigate the GAPDH genes in above species, HMMER v3.0 was employed to perform an HMM search against the established local protein database [40], using the family-specific Gp_dh_N domain (PF00044) and Gp_dh_C domain (PF02800) HMM profiles obtained from the Pfam database (http://pfam.xfam.org/search) [41] as query, with the default parameters and an E-value cutoff of 1e−5. To refine the search results, partial GAPDH domains and other potential false positives were eliminated manually with the Pfam and CDD (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) databases [42]. The information and sequences of Arabidopsis GAPDHs (AtGAPDHs) were retrieved from The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org/).
The previously reported Arabidopsis GAPDH cDNA sequences were used as queries to BLASTn the GenBank EST database for Triticum aestivum (taxid:4565) and Triticum turgidum (taxid:4571). The EST sequences were assembled into contigs using CodonCode Aligner with high stringency parameters of minimum percent identity of 99 %, minimum overlap length of 50 and default parameters for the rest. Open reading frames (ORF) of obtained contigs were carried out with the ORF Finder in NCBI (http://www.ncbi.nlm.nih.gov/gorf/gorf.html), then partial GAPDH domains and other potential false positives were eliminated. Basic data about the GAPDH proteins, such as amino acid number (aa), molecular weight (MW) and isoelectric point (pI), were calculated with ProtParam tool in ExPASy (http://web.expasy.org/protparam/).
Sequence alignment and phylogenetic analysis
Multiple sequence alignment for GAPDHs coding sequences in Arabidopsis, Triticum turgidum, wheat, barley, Aegilops tauschii, and Triticum urartu were performed and edited using MEGA5.1 with the Clustal Omega method [43]. A rooted Neighbor-joining phylogenetic tree of these GAPDHs were constructed with MEGA5.1 under default parameters and bootstrap 1000 [43]. The coding sequences (CDS) and protein sequences of wheat GAPDHs were aligned by DNAMAN 6.0 software with pairwise method [44].
Gene and protein structure analysis
To investigate the exon/intron structures of individual GAPDH gene, we aligned the coding or cDNA sequences with their corresponding genomic DNA sequences. The structure models were collected through the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn) [45]. The GAPDH amino acid sequences were used as input to the MEME website (http://meme-suite.org/tools/meme) to analyze the conserved motifs and plot the motif logos [46].
Expression profile analysis
To analyze the expression profiles of GAPDHs, the microarray data of Arabidopsis thaliana, Hordeum vulgare GAPDHs were downloaded from the BAR and other Data Analysis Tools for Plant Biology (http://bar.utoronto.ca/) with their accession numbers or corresponding probe set IDs and the expression data of wheat GAPDHs were downloaded from PLEXdb (http://www.plexdb.org/) [47]. Combined with the analysis on Genevestigator (https://genevestigator.com/gv/) [48], the tissue expression profiles and the inducible expression profiles in response to stress of GAPDHs were generated by software OriginPro 9.1 and Heatmapper tool in the BAR (http://bar.utoronto.ca/ntools/cgi-bin/ntools_heatmapper.cgi).
Plant materials
Wheat seeds were sterilized with 75 % alcohol and 15 % sodium hypochlorite, rinsed 10 times, and placed on moistened filter paper in Petri dishes and germinated for 1 day and cultivated in a growth room at 22 °C and a 16 h light and 8 h dark photoperiod. Following 10 days of growth, drought and salinity, cold and heat treatments were initiated. The seedlings were grown under 4 or 40 °C for 24 h to simulate cold and heat treatments, respectively; and immersed in 250 mM NaCl or 20 % PEG8000 solutions for 24 h as salt and drought treatments, respectively. Control and treated seeding were harvested for assays at 0, 1, 2, 6, 12 h and 24 h after treatment. All samples were immediately frozen in liquid nitrogen and kept at −80 °C prior to RNA isolation. All experiments were repeated 3 times.
Quantitative real-time PCR (qRT-PCR)
Frozen tissues were ground in liquid nitrogen and total RNA were isolated using the RNAiso plus reagent (TaKaRa, Japan) as per the manufacturer’s specifications and treated with RNase-free DNase I (Invitrogen, USA) for 15 min to degrade any residual genomic DNA. For real-time PCR analysis, first-strand cDNAs were synthesized from DNaseI-treated total RNA using PrimeScript™ RT-PCR Kit (TaKaRa, Japan) according to the manufacturer’s instructions. Real-time PCR was performed in optical 96-well plates (BIOplastics, Netherlands) with CFX96 Touch Real-Time PCR Detection System (BIO-RAD, USA) using the SYBR Green method. The wheat β-actin gene (AB181991) was used as an internal control. The PCR thermal cycle was set up as follows: 95 °C for 1 min; 45 cycles of 94 °C for 15 s, 56 °C for 15 s and 72 °C for 20 s. The quantitative analysis was accomplished with the 2-△△CT method and the relative expression of TaGAPDHs were clustered by MeV v4.9 using the average linkage hierarchical clustering method [49]. The gene specific primers used for quantitative real-time RT–PCR are listed in Additional file 11. Three biological replicates were set up and all experiments were repeated 3 times.
Availability of supporting data
The data sets supporting the results of this article are included within the article and its additional files.
Abbreviations
- GAPDH:
-
glyceraldehyde-3-phosphate dehydrogenase
- G3P:
-
glyceraldehyde-3-phosphate
- GAPA/B:
-
chloroplastic GAPDHs
- GAPC:
-
phosphorylating GAPDH in cytoplasm
- GAPCp:
-
GAPDHs in non-green plastids
- GAPN:
-
cytosolic non-phosphorylating GAPDH
- NtOSAK:
-
tobacco osmotic stress-activated protein kinase
- PLDδ:
-
phospholipase Dδ
- ABA:
-
abscisic acid
- PEG:
-
polyethylene glycerol
References
Sirover MA. On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: biochemical mechanisms and regulatory control. Biochim Biophys Acta. 2011;1810:741–51.
Nicholls C, Li H, Liu JP. GAPDH: a common enzyme with uncommon functions. Clin Exp Pharmacol Physiol. 2012;39:674–9.
Tristan C, Shahani N, Sedlak TW, Sawa A. The diverse functions of GAPDH: views from different subcellular compartments. Cell Signal. 2011;23:317–23.
Kim JW, Dang CV. Multifaceted roles of glycolytic enzymes. Trends Biochem Sci. 2005;30:142–50.
Sirover MA. Subcellular dynamics of multifunctional protein regulation: mechanisms of GAPDH intracellular translocation. J Cell Biochem. 2012;113:2193–200.
Cerff R, Chambers SE. Subunit structure of higher plant glyceraldehyde-3-phosphate dehydrogenases (EC 1.2.1.12 and EC 1.2.1.13). J Biol Chem. 1979;254:6094–8.
Cerff R, Kloppstech K. Structural diversity and differential light control of mRNAs coding for angiosperm glyceraldehyde-3-phosphate dehydrogenases. Proc Natl Acad Sci U S A. 1982;79:7624–8.
Martin W, Cerff R. Prokaryotic features of a nucleus-encoded enzyme. cDNA sequences for chloroplast and cytosolic glyceraldehyde-3-phosphate dehydrogenases from mustard (Sinapis alba). Eur J Biochem. 1986;159:323–31.
Brinkmann H, Cerff R, Salomon M, Soll J. Cloning and sequence analysis of cDNAs encoding the cytosolic precursors of subunits GapA and GapB of chloroplast glyceraldehyde-3-phosphate dehydrogenase from pea and spinach. Plant Mol Biol. 1989;13:81–94.
Velasco R, Salamini F, Bartels D. Dehydration and ABA increase mRNA levels and enzyme activity of cytosolic GAPDH in the resurrection plant Craterostigma plantagineum. Plant Mol Biol. 1994;26:541–6.
Laxalt AM, Cassia RO, Sanllorenti PM, Madrid EA, Andreu AB, et al. Accumulation of cytosolic glyceraldehyde-3-phosphate dehydrogenase RNA under biological stress conditions and elicitor treatments in potato. Plant Mol Biol. 1996;30:961–72.
Wang Q, Kuo L, Sjolund R, Shih MC. Immunolocalization of glyceraldehyde-3-phosphate dehydrogenase in Arabidopsis thaliana. Protoplasma. 1997;198:155–62.
Zaffagnini M, Fermani S, Costa A, Lemaire SD, Trost P. Plant cytoplasmic GAPDH: redox post-translational modifications and moonlighting properties. Front Plant Sci. 2013;4:450.
Petersen J, Brinkmann H, Cerff R. Origin, evolution, and metabolic role of a novel glycolytic GAPDH enzyme recruited by land plant plastids. J Mol Evol. 2003;57:16–26.
Shih MC, Heinrich P, Goodman HM. Cloning and chromosomal mapping of nuclear genes encoding chloroplast and cytosolic glyceraldehyde-3-phosphate-dehydrogenase from Arabidopsis thaliana. Gene. 1991;104:133–8.
Munoz-Bertomeu J, Cascales-Minana B, Mulet JM, Baroja-Fernandez E, Pozueta-Romero J, et al. Plastidial glyceraldehyde-3-phosphate dehydrogenase deficiency leads to altered root development and affects the sugar and amino acid balance in Arabidopsis. Plant Physiol. 2009;151:541–58.
Habenicht A, Hellman U, Cerff R. Non-phosphorylating GAPDH of higher plants is a member of the aldehyde dehydrogenase superfamily with no sequence homology to phosphorylating GAPDH. J Mol Biol. 1994;237:165–71.
Michels S, Scagliarini S, Della Seta F, Carles C, Riva M, et al. Arguments against a close relationship between non-phosphorylating and phosphorylating glyceraldehyde-3-phosphate dehydrogenases. FEBS Lett. 1994;339:97–100.
Wood AJ, Reski R, Frank W. Isolation and characterization of ALDHIIA5, a novel non-phosphorylating GAPDH cDNA from Physcomitrella patens. Bryologist. 2004;107:385–7.
Valverde F, Ortega JM, Losada M, Serrano A. Sugar-mediated transcriptional regulation of the Gap gene system and concerted photosystem II functional modulation in the microalga Scenedesmus vacuolatus. Planta. 2005;221:937–52.
Bustos DM, Bustamante CA, Iglesias AA. Involvement of non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase in response to oxidative stress. J Plant Physiol. 2008;165:456–61.
Morigasaki S, Shimada K, Ikner A, Yanagida M, Shiozaki K. Glycolytic enzyme GAPDH promotes peroxide stress signaling through multistep phosphorelay to a MAPK cascade. Mol Cell. 2008;30:108–13.
Nakamura M, Tomitori H, Suzuki T, Sakamoto A, Terui Y, et al. Inactivation of GAPDH as one mechanism of acrolein toxicity. Biochem Biophys Res Commun. 2013;430:1265–71.
Pelah D, Shoseyov O, Altman A, Bartels D. Water-stress response in Aspen (Populus tremula): differential accumulation of dehydrin, sucrose synthase, GAPDH homologues, and soluble sugars. J Plant Physiol. 1997;151:96–100.
Zhang XH, Rao XL, Shi HT, Li RJ, Lu YT. Overexpression of a cytosolic glyceraldehyde-3-phosphate dehydrogenase gene OsGAPC3 confers salt tolerance in rice. Plant Cell Tissue Organ Cult. 2011;107:1–11.
Wawer I, Bucholc M, Astier J, Anielska-Mazur A, Dahan J, et al. Regulation of Nicotiana tabacum osmotic stress-activated protein kinase and its cellular partner GAPDH by nitric oxide in response to salinity. Biochem J. 2010;429:73–83.
Guo L, Devaiah SP, Narasimhan R, Pan XQ, Zhang YY, et al. Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase D delta to transduce hydrogen peroxide signals in the Arabidopsis response to stress. Plant Cell. 2012;24:2200–12.
Munoz-Bertomeu J, Bermudez MA, Segura J, Ros R. Arabidopsis plants deficient in plastidial glyceraldehyde-3-phosphate dehydrogenase show alterations in abscisic acid (ABA) signal transduction: interaction between ABA and primary metabolism. J Exp Bot. 2011;62:1229–39.
Rius SP, Casati P, Iglesias AA, Gomez-Casati DF. Characterization of an Arabidopsis thaliana mutant lacking a cytosolic non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase. Plant Mol Biol. 2006;61:945–57.
Rius SP, Casati P, Iglesias AA, Gomez-Casati DF. Characterization of Arabidopsis lines deficient in GAPC-1, a cytosolic NAD-dependent glyceraldehyde-3-phosphate dehydrogenase. Plant Physiol. 2008;148:1655–67.
Munoz-Bertomeu J, Cascales-Minana B, Irles-Segura A, Mateu I, Nunes-Nesi A, et al. The plastidial glyceraldehyde-3-phosphate dehydrogenase is critical for viable pollen development in Arabidopsis. Plant Physiol. 2010;152:1830–41.
Chen ZJ, Ha M, Soltis D. Polyploidy: genome obesity and its consequences: Polyploidy workshop: Plant and Animal Genome XV Conference, San Diego, CA, USA, January 2007. New Phytologist. 2007;174:717–20.
Wendel JF. Genome evolution in polyploids. Plant Mol Biol. 2000;42:225–49.
International Wheat Genome Sequencing Consortium (IWGSC). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014;345:1251788.
Marcussen T, Sandve SR, Heier L, Spannagl M, Pfeifer M, et al. Ancient hybridizations among the ancestral genomes of bread wheat. Science. 2014;345.
Manjunath S, Sachs MM. Molecular characterization and promoter analysis of the maize cytosolic glyceraldehyde 3-phosphate dehydrogenase gene family and its expression during anoxia. Plant Mol Biol. 1997;33:97–112.
Jia J, Zhao S, He W, Tao Y, Zhang C, et al. Genomic data from Aegilops tauschii–the progenitor of wheat D genome. GigaScience Database. 2013.
Ling HQ, Zhao S, Zhang C, Tao Y, Gao C, et al. Genomic data from Triticum urartu–the progenitor of wheat A genome. GigaScience Database. 2013.
International Barley Genome Sequencing Consortium. A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012;491:711–6.
Zhang Z, Wood WI. A profile hidden Markov model for signal peptides generated by HMMER. Bioinformatics. 2003;19:307–8.
Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–51.
Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–9.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9.
Kaukonen J, Juselius JK, Tiranti V, Kyttälä A, Zeviani M, et al. Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science. 2000;289:782–5.
Hu B, Jin J, Guo A-Y, Zhang H, Luo J, et al. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2014;31:1296–7.
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, et al. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8.
Dash S, Van Hemert J, Hong L, Wise RP, Dickerson JA. PLEXdb: gene expression resources for plants and plant pathogens. Nucleic Acids Res. 2012;40:D1194–201.
Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, et al. Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics. 2008;2008:420747.
Feng X, Lai Z, Lin Y, Lai G, Lian C. Genome-wide identification and characterization of the superoxide dismutase gene family in Musa acuminata cv. Tianbaojiao (AAA group). BMC Genomics. 2015;16:823.
Acknowledgements
This study was supported by National Natural Science Foundation of China (NO. 31271625).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LZ performed most of the data mining, data analysis, experimental work and illustrations of the figures and tables. RD conceived the study and participated in software implementation and data analysis. ZG helped to conduct qRT-PCR and draft the manuscript. SY and XD gave final approval of the version to be published. All authors read and approved the final manuscript.
Additional files
Additional file 1: Table S1.
HMMsearch hits of wheat Glyceraldehyde 3-phosphate dehydrogenase. (PDF 93 kb)
Additional file 2:
The contigs and singletons of wheat GAPDH that assembled from ESTs. (TXT 704 kb)
Additional file 3:
The contigs and singletons of wheat GAPN that assembled from ESTs. (TXT 9 kb)
Additional file 4:
The GAPDH and GAPN protein sequences of wheat and other species. (TXT 19 kb)
Additional file 5: Table S2.
Pairwise alignments of wheat Glyceraldehyde 3-phosphate dehydrogenase. (PDF 33 kb)
Additional file 6: Table S3.
GAPDH family in plants. (PDF 49 kb)
Additional file 7: Figure S1.
Multiple alignment of wheat GAPDH amino acid sequences. (PDF 462 kb)
Additional file 8: Figure S2.
Multiple alignment of animal/fungi/plant GAPDH amino acid sequences. (PDF 485 kb)
Additional file 9: Figure S3.
Multiple alignment of GPAN amino acid sequences. (PDF 345 kb)
Additional file 10: Figure S4.
Motif LOGOs of GAPDHs and GAPNs generated by MEME. (PDF 593 kb)
Additional file 11: Table S4.
Primers used in this study. (PDF 50 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zeng, L., Deng, R., Guo, Z. et al. Genome-wide identification and characterization of Glyceraldehyde-3-phosphate dehydrogenase genes family in wheat (Triticum aestivum). BMC Genomics 17, 240 (2016). https://doi.org/10.1186/s12864-016-2527-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-016-2527-3