Abstract
In this work, Lactobacillus reuteri has been metabolically engineered for improving 1, 3-propanediol (1, 3-PD) production by the expression of an Escherichia coli alcohol dehydrogenase, yqhD, that is known to efficiently convert the precursor 3-hydroxypropionaldehyde (3-HPA) to 1, 3-PD. The engineered strain exhibited significantly altered formation rates for the product and other metabolites during the fermentation. An increase in the 1, 3-PD specific productivity of 34% and molar yield by 13% was achieved in the clone, relative to the native strain. A concomitant decrease in the levels of toxic intermediate, 3-HPA, was observed, with the specific productivity levels being 25% lesser than that of the native strain. Interestingly, the recombinant strain exhibited elevated rates of lactate and ethanol formation as well as reduced rate of acetate production, compared to the native strain. The preferential utilization of NADPH by YqhD with a possible decrease in the native 1, 3-PD oxidoreductase (NADH-dependent) activity, could have resulted in the diversion of surplus NADH towards increased lactate and ethanol productivities.
Similar content being viewed by others
Introduction
Biological processes are eco-friendly and sustainable alternatives to conventional chemical processes for production of several industrially important bulk chemicals like succinic acid, lactic acid, 1, 3-propanediol, 1, 4-butanediol, etc. (Biebl et al. 1998; Chotani et al. 2000; Song and Lee 2006). Such processes could be economically viable if they are based on renewable feedstocks. Glycerol, a surplus byproduct of the biodiesel industry holds promise as a major feedstock for synthesis of platform chemicals such as 1, 3-propanediol (Zhu et al. 2002). Currently, 1, 3-propanediol (1, 3-PD) has attracted worldwide interest due to its enormous applications in polymers, cosmetics, foods, adhesives, lubricants, laminates, solvents, antifreeze and medicines (Homann et al. 1990; Colin et al. 2000; Zhu et al. 2002; Cheng et al. 2007).
The biological route involves 1, 3-PD production by microorganisms like Klebsiella, Citrobacter, Enterobacter, Clostridia and Lactobacilli (Biebl et al. 1999; Saxena et al. 2009). Amongst these, Clostridium butyricum and Klebsiella pneumoniae, are considered to be the best producers (Gonzalez-Pajuelo et al. 2006). 1, 3-PD concentrations in the range of around 40 - 100 g/l have been obtained with these producers (Celinska 2010). The product levels of the native producers have been improved using various bioprocess strategies. Metabolic engineering is currently being attempted to further enhance the product levels (Saxena et al. 2009).
The non-native producers, Escherichia coli and Saccharomyces cerevisiae, have also been engineered for 1, 3-PD production. In S. cerevisiae, due to ineffective transport of vitamin B12 needed for 1, 3-PD synthesis, only low levels of the product has been obtained. On the other hand, E. coli has been metabolically engineered by DuPont and Genencor International, Inc., to produce 1, 3-PD at a concentration of 135 g/l, (Maervoet et al. 2011) the highest reported so far in the industry. A major concern with the existing 1, 3-PD producers is that a majority of them are opportunistic pathogens, that are less suitable for niche applications in food, cosmetic and biomedical industries. In this context Lactobacillus reuteri, a GRAS (generally regarded as safe) organism, offers immense potential as a host for 1, 3-PD production.
Lactobacillus reuteri converts glycerol to 1, 3-PD in a two-step anaerobic process (Figure 1). In the first step, a cobalamin-dependent glycerol dehydratase catalyzes the conversion of glycerol to 3-hydroxypropionaldehyde (3-HPA). In the second step, 3-HPA is reduced to 1, 3-PD by a NADH-dependent oxidoreductase (Talarico et al. 1990). 1, 3-PD productivity of around 10-30 g/l has been achieved so far in native L. reuteri (Baeza-Jimenez et al. 2011; Tobajas et al. 2009).
Pathways of glucose and glycerol metabolism in L. reuteri. Abbreviations: G6P glucose-6-phosphate, 6PG 6-phosphogluconate, R5P ribulose-5-phosphate, X5P xylulose-5-phosphate, AcP acetyl phosphate, AcCoA acetyl-CoA, F6P fructose-6-phosphate, FBP, fructose-1, 6-bisphosphate, DHAP dihydroxyacetone phosphate, GAP glyceraldehyde-3-phosphate, Pyr pyruvate, G3P glycerol-3-phosphate, 3-HPA 3-hydroxypropionaldehyde, GDHt glycerol dehydratase, 1, 3-PDOR 1, 3-propanediol oxidoreductase in L. reuteri, YqhD E. coli alcohol dehydrogenase.
The major bottleneck limiting 1, 3-PD production in L. reuteri is growth inhibition by secreted metabolites and toxic 3-HPA. These metabolites are produced to regenerate the cofactors such as NADH/NADPH. Therefore redirecting flux from these competing pathways towards product formation by balancing the redox potential would be a powerful metabolic engineering strategy. For instance, disruption of ethanol synthesis has been demonstrated to substantially improve flux through the 1, 3-PD biosynthetic pathway in K. pneumoniae (Zhang et al. 2006). Further, redirection of flux from central carbon metabolism towards 1, 3-PD synthesis should be complemented by adequate levels of enzymes and cofactors involved in the pathway.
In this work, we have expressed an E. coli alcohol dehydrogenase, yqhD, in L. reuteri, to increase 1, 3-PD productivity by improved conversion of 3-HPA. Further, the impact of the heterologously expressed yqhD on cell growth, 1, 3-PD production and byproduct formation has been analyzed.
Materials and methods
Strains and plasmids
The bacterial strains and plasmids used and modified in this study are listed in Table 1.
Media and growth conditions
L. reuteri ATCC 55730 and the E. coli strains were grown at 37°C in MRS (MRS contains 5 g yeast extract, 10 g proteose peptone, 10 g beef extract, 2 g dipotassium phosphate, 2 g ammonium citrate, 5 g sodium acetate, 100 mg magnesium sulphate, 50 mg manganese sulphate, 1 g polysorbate 80 and 20 g dextrose, per liter) broth and LB broth, respectively. The recombinants were cultured in media containing appropriate antibiotics, ampicillin (100 μg/mL) and erythromycin (200 μg/mL for E. coli and 5 μg/mL for L. reuteri). Growth was monitored by measuring the absorbance at 600 nm. Cell dry weight (CDW) was calculated from a predetermined relationship between L. reuteri CDW and optical density (1 OD600 corresponded to 0.33 g/l CDW).
Chemicals and Reagents
The enzymes and reagents used in cloning experiments - Nco I, Xho I, T4 DNA ligase, and Phusion™ Flash High-Fidelity PCR Master Mix - were bought from New England Biolabs (Manassas, USA). Plasmid miniprep spin kit and PCR purification kit were procured from Qiagen (Germany). Primers were procured from VBC-Biotech (Austria) and the inducer sakacin P induction peptide (SppIP) was synthesized from GenScript (USA). Culture media (LB and MRS), the antibiotics, erythromycin and ampillicin, and other chemicals were purchased from HiMedia Laboratories (Mumbai, India). Since 3-HPA standard could not be commercially procured, it was synthesized in our laboratory as described under "3-HPA production by resting cells of L. reuteri ATCC 55730".
Construction of the recombinant plasmids
A schematic representation of the structure of recombinant plasmid, pHR2, carrying yqhD, is shown in Figure 2. The 1.163 kb yqhD gene fragment (GenBank accession number NC010498), was amplified from the chromosomal DNA of E. coli K-12 MG1655 using the primers yqhDF and yqhDR (Table 2). PCR conditions employed were - an initial denaturation at 98°C (10 s), followed by 25 cycles of the program: 98°C (3 s); 65°C (5 s); 72°C (20 s) and a final extension at 72°C (1 min).
Structure of the recombinant plasmid pHR2 (~6.86 kb). yqhD E. coli alcohol dehydrogenase gene, open rectangle MCS, TpepN transcription terminator, sh71rep replication origin for Lactobacillus, ermL erythromycin-resistance marker, PssIP and PorfX inducible promoters, sppK and sppR histidine protein kinase and response regulator respectively.
The amplicon was cloned into TA vector to generate the recombinant plasmid pHR1. Further, the yqhD gene was sub-cloned from pHR1 into Nco I/Xho I site of pSIP411, resulting in recombinant plasmid, pHR2. The clones were screened by lysate PCR using the primer pair PorfXF and yqhDR (Table 2). The plasmid pHR2 was electroporated into L. reuteri to yield, L. reuteri HR2. The electrocompetent cells were prepared as described by Berthier et al. (1996). Electroporation was performed with a BTX electroporator, using pulse settings of 1.5 kV, 800 Ω and 25 μF and a time constant of 11 - 13 ms was obtained. The cells were plated on MRS agar containing the required antibiotic and incubated for 24 - 36 h at 37°C until visible colonies were observed. The recombinant plasmid pHR2 was isolated from L. reuteri HR2 using the plasmid miniprep kit, with the following modifications: The cells in resuspension buffer, were lysed with 30 mg/mL lysozyme (USB) and incubated at 37°C for 30 minutes. The rest of the procedure was as per the miniprep manual (Qiagen).
Batch fermentation
The inoculum for the batch reactor was grown in 150 mL MRS broth with erythromycin at 37°C until an OD600 of 0.8 - 1.0 was reached. The seed was then inoculated into a 2 L fermentor (KLF 2000 - Bioengineering AG, Switzerland) filled with 1.2 L MRS medium containing erythromycin and glycerol (278 mM). A glucose to glycerol ratio of 1:2.5 has been used in this study for elevated 1, 3-PD synthesis (Tobajas et al. 2009). Fermentation was carried out at 37°C and 250 rpm, in an anaerobic condition. The pH was maintained at 5.5 by the addition of 1.5 M NaOH or 1.5 M H3PO4 (El-Ziney et al. 1998). The anaerobic condition was established by flushing with sterile nitrogen. At 0.8 OD600, the culture was induced with 50 ng/mL of sakacin P induction peptide (SppIP). Samples were removed periodically for determining OD600. The culture pellet and supernatant were stored at - 20°C, to be used later for protein and metabolite analyses respectively.
Substrate and Metabolite Analyses
Concentrations of glucose, glycerol, 1, 3-PD, ethanol, lactate, 3-HPA and acetate in the culture broth were determined using a HPLC (Shimadzu LC-10AT VP) that was equipped with a refractive index detector (RID) and an aminex HPX-87H column (300 × 78 mm, Bio-Rad, USA). The mobile phase consisted of acetonitrile and water in a ratio of 35:65 in 5 mM H2SO4, at 0.4 mL/min. The temperature of column and RID was maintained at 30°C and 50°C respectively. Samples were filtered through 0.22 μm filters before analysis. 3-HPA standard was synthesized in the lab using resting cells of L. reuteri ATCC 55730 as explained below. Quantitation of 3-HPA was done by HPLC, as described by Spinler et al. (2008).
3-HPA production by resting cells of L. reuteri ATCC 55730
3-HPA was produced as described previously (Spinler et al. 2008; Luthi-Peng et al. 2002). Briefly, L. reuteri was cultured in 100 mL MRS broth, incubated anaerobically at 37°C for 24 h. The anaerobic condition was maintained by sparging with nitrogen. The culture was centrifuged and the pellet washed with 50 mM sodium phosphate buffer (pH 7.4). The cells were resuspended in 250 mM glycerol to a concentration of ~1.5 × 1010 cells/mL and incubated anaerobically at 37°C for 2 h. After the 2 h incubation, the culture was pelleted and the 3-HPA-containing supernatant was collected and filter-sterilized using a 0.22 μm filter and the filtrate used for HPLC analysis.
SDS-PAGE analysis of yqhD expression in L. reuteri
The SDS-PAGE was conducted on a 12% polyacrylamide gel (Laemmli 1970). The proteins on the gel were stained with 0.025% (w/v) Coomassie Brilliant Blue G-250. Protein concentration was determined by the Bradford method (Bradford 1976) with bovine serum albumin (BSA) as standard.
Results
Heterologous expression of alcohol dehydrogenase (yqhD) in Lactobacillus reuteri ATCC 55730
The E. coli alcohol dehydrogenase gene (yqhD) was cloned and expressed in L. reuteri. The recombinant plasmid, pHR2 with yqhD gene was constructed as shown in Figure 2. The expression of the cloned yqhD gene in L. reuteri was confirmed using SDS-PAGE analysis of whole cell lysates (Figure 3). A prominent band of ~43 kDa appeared in the recombinant cells after induction, which correlates well with the expected size of YqhD.
Batch fermentation analysis of recombinant L. reuteri harbouring yqhD
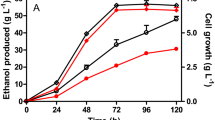
To investigate the impact of yqhD expression on cell growth, substrate consumption, formation of 1, 3-PD, 3-HPA and other metabolites, batch fermentation of recombinant L. reuteri was carried out, with native strain as control. The cell concentration of both native and recombinant strains reached around 1.8 and 1.4 g/l of CDW respectively. The specific growth rate (μmax) of the recombinant strain was lower (0.38 h-1) compared to the wild type (0.46 h-1) (Figure 4).
It was observed that yqhD expression in L. reuteri, altered the specific substrate uptake, product and byproduct formation rates significantly (Figure 5). The specific production rate of 1.38 g/g h for 1, 3-PD in the recombinant strain achieved during the log phase after induction, was notably higher (by 34%) than that of the native strain (1.03 g/g h) (Figure 5). This correlates with a 25% decrease in the levels of 3-HPA secreted in the recombinant culture (0.14 g/g h), relative to the native strain (0.19 g/g h) (Figure 5). This enhanced 3-HPA conversion has supposedly contributed to the increased molar yield of 1, 3-PD (up by 13%) observed in the clone (Table 3). Interestingly, the specific rates of formation of lactate and ethanol were higher and that of acetate lower in the recombinant culture, relative to the native strain, during the second half of the logarithmic phase (Figure 5).
The batch experiment has revealed that 1, 3-PD, acetate and ethanol are growth-associated in both the native and recombinant L. reuteri strains, while lactate and 3-HPA are growth-associated only in the recombinant strain (Figure 6a, b). During the glucose-glycerol cofermentation, consumption of these two carbon sources was not synchronous. Glucose was consumed more rapidly than glycerol during the early log phase and was exhausted before glycerol in both the native and recombinant strains (Figure 4). In the recombinant strain, glycerol is not utilized upon exhaustion of glucose, while the native strain exhibited moderate glycerol consumption and concomitant 3-HPA synthesis even after depletion of glucose (Figure 4, 6b). However, 1, 3-PD synthesis is observed only when both the carbon sources are utilized in the recombinant and in the native strains during the late-log and early-stationary phase (Figure 4, 6b)
Discussion
L. reuteri produces 1, 3-PD along with 3-HPA only when glycerol is cofermented with glucose. Lower glucose levels have been shown to favour 3-HPA formation. Higher glucose concentrations generate more NADH, that is consumed for reducing 3-HPA to 1, 3-PD. Glycerol serves as an electron sink by recycling NADH produced during glycolysis (Luthi-Peng et al. 2002; Schutz and Radler 1984). In this work, 1, 3-PD synthesis is observed both in native and recombinant strains only when both the carbon sources are utilized (Figure 4, 6b). In the case of native strain, glycerol consumption upon exhaustion of glucose resulted in 3-HPA accumulation, since NADH supply could be limited by reduced glycolysis. Thus redox balance plays a crucial role in 1, 3-PD formation.
Enhancing the enzyme concentration and cofactor availability could lead to improved 1, 3-PD formation. As the phosphoketolase pathway prevalent in L. reuteri (Årsköld et al. 2008), provides increased NADPH, overexpression of yqhD, has the potential to further improve 1, 3-PD productivity. In this work, expression of yqhD has increased the molar yield of 1, 3-PD from glycerol by 13% in L. reuteri HR2. This is in contrast to the results reported by Zhuge et al. (2010) in recombinant K. pneumoniae strain, wherein yqhD overexpression did not increase the 1, 3-PD yield. However, upon overexpression of yqhD, they have observed a reduction in the activity of the native 1, 3-PD oxidoreductase (1, 3 PDOR), with increased ethanol production. A similar diminishing activity of the native 1, 3 PDOR is perceived in L. reuteri HR2, along with elevated rates of lactate and ethanol production.
The enhanced formation rates of lactate and ethanol observed in the recombinant L. reuteri strain could be indirectly linked to the preferential utilization of NADPH by YqhD for 3-HPA conversion. The consumption of NADPH by YqhD and a possible reduction in the native NADH-dependent 1, 3-PDOR activity could have led to an increased cellular NADH/NAD+ ratio. The surplus NADH thus generated has been diverted for the production of NADH-consuming metabolites like lactate and ethanol.
The elevated specific production rate of ethanol with concomitant decrease in specific acetate production rate implies that acetyl phosphate is channeled more towards ethanol production (Figure 5). This is most likely reflected as a shift in metabolism from acetate to ethanol production, resulting in reduced ATP synthesis. The decreased ATP production coupled with the diversion of NADPH away from biosynthesis by YqhD, could have contributed to the decreased growth rate of the recombinant culture (Jarboe et al. 2010; Zhu et al. 2009). The decreased μmax of the recombinant strain could also be attributed to the metabolic load imposed by the recombinant plasmid on the host (Bentley et al. 1990). Further, metabolic flux analysis needs to be carried out by measuring the enzyme activities and cofactors to verify this hypothesis. The present work has indicated that metabolic engineering can be effectively used to enhance 1, 3-PD productivity in L. reuteri. Further engineering of the strain to improve the redox balance and minimize the formation of byproducts like lactate and ethanol could pave the way for maximizing 1, 3-PD biosynthesis.
References
Årsköld E, Lohmeier-Vogel EM, Cao R, Roos S, Rådström P, van Niel EWJ: Phosphoketolase pathway dominates in Lactobacillus reuteri ATCC 55730 containing dual pathways for glycolysis. J Bacteriol 2008, 190: 206–212. 10.1128/JB.01227-07
Baeza-Jiménez R, López- Martinez LX, De la Cruz-Medina J, Espinosa-de-los-Monteros JJ, García- Galindo HS: Effect of glucose on 1, 3-propanediol production by Lactobacillus reuteri . Revista Mexicana de Ingenierίa Quίmica 2011, 10: 39–46.
Bentley WE, Mirjalili N, Andersen DC, Davis RH, Kompala DS: Plasmid-encoded protein: The principal factor in the "metabolic burden" associated with recombinant bacteria. Biotechnol Bioeng 1990, 35: 668–681. 10.1002/bit.260350704
Berthier F, Zagorec M, Champomier-Verge's M, Ehrlich SD, Morel-Deville F: Efficient transformation of Lactobacillus sake by electroporation. Microbiology 1996, 142: 1273–1279. 10.1099/13500872-142-5-1273
Biebl H, Menzel K, Zeng AP, Deckwer WD: Microbial production of 1, 3-propanediol. Appl Microbiol Biotechnol 1999, 52: 289–297. 10.1007/s002530051523
Biebl H, Zeng AP, Menzel K, Deckwer WD: Fermentation of glycerol to 1, 3-propanediol and 2, 3-butanediol by Klebsiella pneumoniae . Appl Microbiol Biotechnol 1998, 50: 453–457.
Bradford MM: Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976, 72: 248–254. 10.1016/0003-2697(76)90527-3
Celińska E: Debottlenecking the 1, 3-propanediol pathway by metabolic engineering. Biotechnol Adv 2010, 28: 519–530. 10.1016/j.biotechadv.2010.03.003
Cheng KK, Zhang JA, Liu DH, Sun Y, Liu HJ, Yang MD: Pilot-scale production of 1, 3-propanediol using Klebsiella pneumoniae . Process Biochem 2007, 42: 740–744. 10.1016/j.procbio.2007.01.001
Chotani G, Dogde T, Hsu A, Kumar M, LaDuca R, Trimbur D, Weyler W, Sanford K: The commercial production of chemicals using pathway engineering. Biochim Biophys Acta 2000, 1543: 434–55. 10.1016/S0167-4838(00)00234-X
Colin T, Bories A, Moulin G: Inhibition of Clostridium butyricum by 1, 3-propanediol and diols during glycerol fermentation. Appl Microbiol Biotechnol 2000, 54: 201–205. 10.1007/s002530000365
El-Ziney MG, Arneborg N, Uyttendaele M, Debevere J, Jakobsen M: Characterization of growth and metabolite production of Lactobacillus reuteri during glucose/glycerol cofermentation in batch and continuous cultures. Biotechnol Lett 1998, 20: 913–916. 10.1023/A:1005434316757
Gonzalez-Pajuelo M, Meynial-Salles I, Mendes F, Soucaille P, Vasconcelos I: Microbial conversion of glycerol to 1, 3-propanediol: physiological comparison of a natural producer, Clostridium butyricum VPI 3266, and an engineered strain, Clostridium acetobutylicum DGI(pSPD5). Appl Environ Microbiol 2006,72(1):96–101. 10.1128/AEM.72.1.96-101.2006
Homann T, Tag C, Biebl H, Deckwer WD, Schink B: Fermentation of glycerol to 1, 3-propanediol by Klebsiella and Citrobacter strains. Appl Microbiol Biotechnol 1990, 33: 121–126.
Jarboe LR: YqhD: a broad-substrate range aldehyde reductase with various applications in production of biorenewable fuels and chemicals. Appl Microbiol Biotechnol 2010, 10: 2912–2919.
Laemmli UK: Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227: 680–685. 10.1038/227680a0
Lüthi-Peng Q, Dileme FB, Puhan Z: Effect of glucose on glycerol bioconversion by Lactobacillus reuteri . Appl Microbiol Biotechnol 2002, 59: 289–296. 10.1007/s00253-002-1002-z
Maervoet VET, De Mey M, Beauprez J, De Maeseneire S, Soetaert WK: Enhancing the microbial conversion of glycerol to 1, 3-Propanediol using metabolic engineering. Org Process Res Dev 2011, 15: 189–202. 10.1021/op1001929
Saxena RK, Anand P, Saran S, Isar J: Microbial production of 1, 3-propanediol: recent developments and emerging opportunities. Biotechnol Adv 2009, 27: 895–913. 10.1016/j.biotechadv.2009.07.003
Schutz H, Radler F: Anaerobic reduction of glycerol to 1, 3-propanediol by Lactobacillus brevis and Lactobacillus buchneri . Syst Appl Microbiol 1984, 5: 169–178.
Song H, Lee SY: Production of succinic acid by bacterial fermentation. Enzyme Microb Technol 2006, 39: 352–361. 10.1016/j.enzmictec.2005.11.043
Sørvig E, Mathiesen G, Naterstad K, Eijsink VGH, Axelsson L: High-level, inducible gene expression in Lactobacillus sakei and Lactobacillus plantarum using versatile expression vectors. Microbiology 2005, 151: 2439–2449. 10.1099/mic.0.28084-0
Spinler JK, Taweechotipatr M, Rognerud CL, Ou CN, Tumwasorn S, Versalovic J: Human-derived probiotic Lactobacillus reuteri demonstrate antimicrobial activities targeting diverse enteric bacterial pathogens. Anaerobe 2008, 14: 166–171. 10.1016/j.anaerobe.2008.02.001
Talarico TL, Axelsson LT, Novotny J, Fiuzat M, Dobrogosz WJ: Utilization of glycerol as a hydrogen acceptor by Lactobacillus reuteri : purification of 1, 3-propanediol:NAD+ oxidoreductase. Appl Environ Microbiol 1990, 56: 1195–7.
Tobajas M, Mohedano AF, Casas JA, Rodriguez JJ: Unstructured kinetic model for reuterin and 1, 3-propanediol production by Lactobacillus reuteri from glycerol/glucose cofermentation. J Chem Technol Biotechnol 2009, 84: 675–680. 10.1002/jctb.2098
Zhang Z, Li Y, Du C, Liu M, Cao Z: Inactivation of aldehyde dehydrogenase: a key factor for engineering 1, 3-propanediol production by Klebsiella pneumoniae . Metab Eng 2006, 8: 578–586. 10.1016/j.ymben.2006.05.008
Zhu JG, Li S, Ji XJ, Huang H, Hu N: Enhanced 1, 3-propanediol production in recombinant Klebsiella pneumoniae carrying the gene yqhD encoding 1, 3-propanediol oxidoreductase isoenzyme. World J Microbiol Biotechnol 2009, 25: 1217–1223. 10.1007/s11274-009-0005-7
Zhu MM, Lawman PD, Cameron DC: Improving 1, 3-propanediol production from glycerol in a metabolically engineered Escherichia coli by reducing accumulation of sn-glycerol-3-phosphate. Biotechnol Prog 2002, 18: 694–699. 10.1021/bp020281+
Zhuge B, Zhang C, Fang H, Zhuge J, Permaul K: Expression of 1, 3-propanediol oxidoreductase and its isoenzyme in Klebsiella pneumoniae for bioconversion of glycerol into 1, 3-propanediol. Appl Microbiol Biotechnol 2010, 87: 2177–2184. 10.1007/s00253-010-2678-0
Acknowledgements
This work was supported by the grant (No. SR/SO/BB-39/2008) from Department of Science and Technology, New Delhi-110 016, India. Partial grant of fellowship from the Department of Biotechnology, Government of India, is duly acknowledged. We thank DIC at the Centre for Biotechnology, Anna University for providing computational facility. We also gratefully acknowledge Biogaia AB, Sweden, for kindly providing us Lactobacillus reuteri ATCC 55730, Dr Jan Kok for E. coli EC 1000 strain, Dr Takashi Horiuchi for E. coli K-12 strain and Dr Lars Axelsson for pSIP411 vector. We thank our colleague Mr K Chandru (Centre for Biotechnology, Anna University, Chennai, India), for assisting with protein expression analysis.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Vaidyanathan, H., Kandasamy, V., Gopal Ramakrishnan, G. et al. Glycerol conversion to 1, 3-Propanediol is enhanced by the expression of a heterologous alcohol dehydrogenase gene in Lactobacillus reuteri . AMB Expr 1, 37 (2011). https://doi.org/10.1186/2191-0855-1-37
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/2191-0855-1-37