Abstract
Previously, we explored the epidemic pattern and molecular characterization of enteroviruses isolated in Chungnam, Korea from 2005 to 2006. The present study extended these observations to 2008 and 2009. In this study, enteroviruses showed similar seasonal prevalent pattern from summer to fall and age distribution to previous investigation. The most prevalent month was July: 42.9% in 2008 and 31.9% in 2009. The highest rate of enterovirus-positive samples occurred in children < 1-year-old-age. Enterovirus-positive samples were subjected to sequence determination of the VP1 region, which resolved the isolated enteroviruses into 10 types in 2008 (coxsackievirus A4, A16, B1, B3, echovirus 6, 7, 9, 11, 16, and 30) and 8 types in 2009 (coxsackievirus A2, A4, A5, A16, B1, B5, echovirus 11, and enterovirus 71). The most prevalent enterovirus serotype in 2008 and 2009 was echovirus 30 and coxsackievirus B1, respectively, whereas echovirus 18 and echovirus 5 were the most prevalent types in 2005 and 2006, respectively. Comparison of coxsackievirus B1 and B5 of prevalent enterovirus type in Korea in 2009 with reference strains of each same serotype were conducted to genetic analysis by a phylogenetic tree. The sequences of coxsackievirus B1 strains segregated into four distinct clusters (A, B, C, and D) with some temporal and regional sub-clustering. Most of Korean coxsackievirus B1 strains in 2008 and 2009 were in cluster D, while only "Kor08-CVB1-001CN" was cluster C. The coxsackievirus B5 strains segregated in five distinct genetic groups (clusters A-E) were supported by high bootstrap values. The Korean strains isolated in 2001 belonged to cluster D, whereas Korean strains isolated in 2005 and 2009 belonged to cluster E. Comparison of the VP1 amino acid sequences of the Korean coxsackievirus B5 isolates with reference strains revealed amino acid sequence substitutions at nine amino acid sequences (532, 562, 570, 571, 576-578, 582, 583, and 585).
Similar content being viewed by others
Introduction
Human enteroviruses (HEV) are RNA viruses from the picornaviridae family. Clinical consequences of HEV infections include the common cold, hand-foot-mouth disease, acute hemorrhagic conjunctivitis, myocarditis, encephalitis, poliomyelitis, and aseptic meningitis. The latter, which mainly affects young children, is the most commonly encountered illness associated with enteroviral infections, often appearing in the form of outbreaks [1–3]. More than 80 immunologically distinct serotypes cause infections in humans; they are grouped into polioviruses, echoviruses (ECV), coxsackievirus A (CVA), coxsackievirus (CVB), and enterovirus (EV) types 68-71. These viruses are also divided into several subgroups: polioviruses and HEV-A, HEV-B, HEV-C, and HEV-D [4–7].
Outbreaks of enteroviruses typically peak during the summer and early fall, and various serotypes are often associated with a single outbreak. The predominant enterovirus types vary from year-to-year, with ECV 9, 13, 18, and 30 and CVB 5 being the most frequently isolated in Europe and the United States over the past few years [8–10]. Since 1993, when nationwide surveillance began in Korea, there had been reports of summer outbreaks of enteroviruses, involving ECV 5, 6, 7, 9, 13, 18, and 30, CVA 24, CVB 3 and 5, and EV 71 [11, 12].
The enterovirus genome contains a 7,500 nucleotide-long single-stranded RNA molecule with polarity. The 5' and 3' non-coding regions (NCRs) are generally highly conserved in general. The most variable regions of the genome are within the genes encoding the capsid proteins, VP1, VP2, VP3 and VP4, which are partially exposed on the virus surface [13–15]. Laboratory diagnosis of enterovirus infections is based on amplification of highly conserved regions within the enteroviral RNA genome, The 5'NCR seems to be the most conserved region among enteroviruses and is therefore targeted widely in diagnostic procedures [16, 17]. In addition to traditional virological methods used to identify the enterovirus serotype, reverse transcription-polymerase chain reaction (RT-PCR) methods based on amplification of VP1 region have been recently developed [18–20]. Since the VP1 region is one of the main exposed regions of the viral capsid and has been suggested to include a serotype specific antigenic neutralization site, the BC loop in the VP1 region is, in particular, one of the regions associated with viral antigenicity, and substitutions resulting in conformational changes in this region are believed to play a role in host adaptation for enteroviruses [21–23]. Appropriately, the partial VP1 sequences were compared with a database of complete enterovirus VP1 sequences of all serotypes to determine whether the isolates were related genetically to any known enterovirus serotype [19]. In addition, phylogenetic analysis from sequence data of the VP1 region is considered to be a standard method of molecular analysis for epidemiological purposes and the clustering or genotyping in combination with phylogenetic analysis is able to discriminate between lineages within a serotype and to identify emerging new variants or serotypes [24].
An epidemic due to enterovirus occurred in Chungnam, Korea in 2008 and 2009. Presently, to determine the enterovirus serotypes of the epidemic, viral cultures were carried out by inoculating samples to susceptible cell lines and examining the cytopathic effects. Also, molecular detection was performed using 5' NCR RT-PCR and sequencing of the VP1 region of enteroviruses. The aim was to determine the epidemiology of the enterovirus infection and molecular characteristics of the Korean CVB 1 and CVB 5 isolates.
Materials and methods
Virus isolation
Using susceptible cell lines such as rhabdomyosarcoma (Rd), Vero, and buffalo green monkey (BGM) cells, enteroviruses were isolated from 1,214 clinical stool or cerebrospinal fluid (CSF) specimens from hospitalized patients whose symptoms were consistent with enterovirus infections in Chungnam, Korea in 2008 and 2009.
RT-PCR
The testing algorithm for detection and molecular typing of enterovirus was previously described [11]. Cells exhibiting 70% cytopathic effects were frozen and thawed three times and viral RNA was extracted from the supernatant of the infected cells using Magnetic-beads (Toyobo, Osaka, Japan). The extracted RNA was dissolved in 50 μL of nuclease-free water and stored at -70°C until used for RT-PCR. For cDNA synthesis, a 20 μL reaction mixture containing 5 μL of each viral RNA, 0.2 μL primer (AN32, AN33, AN34, and AN35) (Table 1), 4 μL of 5X reverse transcriptase buffer, 2 μL of 0.1 M dithiothreitol (DTT), 4 μL of 10 mM M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA) was used. The mixture was reacted at 20°C for 10 min, 37°C for 120 min, 95°C for 5 min, and then chilled on ice. PCR was performed using a primer set specific for the 5' NCR of the enterovirus as described previously [25]. Briefly, a 50 μL reaction mix containing 0.2 μM of primers ENT-F and ENT-R (Table 1), 2U of Taq DNA polymerase (Promega, Madison, WI), 100 μM concentrations of mixture of dNTPs, and 2 μM MgCl2 was amplified by 35 cycles of 94°C for 1 min, 52°C for 1 min, and 72°C for 1 min. The final extension step was extended to 72°C for 7 min. Semi-nested PCR amplifying the VP1 coding region was carried out as described previously [18]. In the initial PCR, a 50 μL reaction mix containing 0.2 μM of primers 224 and 222 (Table 1), 2 U of Taq DNA polymerase (Promega), 100 μM concentrations of mixture of dNTPs, a 2 μM MgCl2 was amplified by 40 cycles of 95°C for 30 sec, 42°C for 30 sec, and 60°C for 45 sec. One microliter of the first PCR product was added to a second PCR for semi-nested amplification. Fifty microliters of a reaction mix containing 0.2 μM of primers AN89 and AN88 (Table 1), 2.5 U of Taq DNA polymerase (Promega), 100 μM concentrations of a mixture of dNTPs, and 2 μM MgCl2 was incubated at 95°C for 6 min prior to 40 amplification cycles of 95°C for 30 sec, 60°C for 20 sec, and 72°C for 15 sec.
Nucleotide sequencing and molecular typing
PCR products were purified using the QIA quick PCR purification kit (Qiagen, Valencia, CA). Purified DNA was added in a reaction mixture containing 2 μL of Big Dye terminator reaction mix (Applied Biosystems, Foster City, CA) and 2 pmoles of AN88 and AN89 primers (Table 1). Sequencing reactions were subjected to initial denaturation at 96°C for 1 min and 25 cycles consisting of 96°C for 10 sec, 50°C for 5 sec, and 60°C for 4 min in a Gene Amp PCR system 2700 (Applied Biosystems). The products were purified by precipitation with 100% cold ethanol and 3 M sodium-acetate (pH 5.8), and then loaded on a model 3100 automated genetic analyzer (Applied Biosystems). The molecular type of each isolates was determined by the serotype of the highest scoring strain in Genbank using the Basic Local Alignment Search Tool (BLAST); that is, the sequence of the enterovirus strain that gave the highest nucleotide similarity value with the query sequence [26].
Sequence analysis of CVB 1 and CVB 5
Seventeen Korean CVB 1 and eight Korean CVB 5 strains that displayed a more informed profile were selected for sequence analysis (Table 1). Nucleotide and deduced amino acid sequences of candidate enterovirus isolates were compared with the reference strains using CLUSTAL W (version 1.81) and Megalign (DNASTAR) [27]. These programs are applied Needleman and Wunsch algorithm (M-NW similarity test). A similarity score between each pair of sequences was obtained manually after sequential pairwise alignment (M-NW similarity test) was performed. The quality of the alignment could not be measured, and the gap opening penalty (GOP) and the gap extension penalty (GEP) values were used as default values [28, 29]. The phylogenetic relationships among the VP1 sequences of each virus isolate were inferred by using MEGA software v. 4.0. Maximum Composite Likelihood was used as the substitution method, while the neighbor-joining method was used to reconstruct the phylogenetic tree [30]. The reliability of the phylogenetic tree was determined by bootstrap re-sampling of 1,000 replicates.
Nucleotide sequence accession numbers
The enterovirus candidates sequences reported here were deposited in the Genbank sequence database, with the accession numbers summarized in Table 1.
Results
Enterovirus detection and molecular typing
In 2008 and 2009, 1214 samples obtained from patients with aseptic meningitis and enterovirus-related disease were subjected to a diagnostic RT-PCR with cell culture that generated 436 bp amplicons, corresponding to a highly conserved domain in 5' NCR. Eighty two enteroviruses were isolated from 435 cases (18.9%) in 2008 and 107 enteroviruses were isolated from 779 cases (13.7%) in 2009. For molecular typing and phylogenetic analysis, the VP1 amplicons generated in the semi-nested PCR were sequenced and were determined to correspond with a 372 bp VP1 region. Gapped BLAST analyses were carried out and each virus was assigned the type that gave the highest VP1 identity score. The types in the 189 isolates were identified as ECV 30 (n = 42, 22.2%), CVB 1 (n = 37, 19.6%), CVB 5 (n = 21, 11.1%), ECV 6 (n = 19, 10.1%), EV 71 (n = 12, 6.3%), CVA 2 (n = 11, 5.8%), CVA 16 (n = 9, 4.8%), CVA 4 (n = 6, 3.2%), CVA 5 (n = 3, 1.6%), ECV 5 (n = 3, 1.6%), ECV 11 (n = 3, 1.6%), CVB 3 (n = 2, 1.1%), ECV 9 (n = 2, 1.1%), and ECV 16 (n = 1, 0.5%). VP1 amplicons were not generated from 18 samples by semi-nested PCR. (Table 2)
Epidemiological features of enterovirus in Chungnam
Temporal distribution of the epidemic enterovirus in Chungnam was seasonal, with most cases occurring during the summer from June to September. The enterovirus detection rate in these months in 2008 and 2009, respectively, was 25.0% and 16.7% in June, 42.9% and 31.9% in July, 39.2% and 26.4% in August, and 14.3% and 17.0% in September (Figure 1).
Seventy four cases (90.2%) of 82 total samples in 2008 and 102 cases (95.3%) of 107 total cases in 2009 involved individuals < 10-years-of-age. Eight cases (9.8%) in 2008 and five cases (4.7%) in 2009 involved individuals > 10-years-of-age. The highest rate of enterovirus-positive samples were from patients < 1-year-of-age (31.7% in 2008 and 57.0% in 2009) (Figure 2.). Of the total 189 isolates, 121 were from males and 68 were from females, giving a male-to-female ratio of approximately 1.78:1.
The major clinical symptoms of hospitalized patients infected by enterovirus are summarized in Table 3. Aseptic meningitis was the major clinical manifestation (80.5% in 2008 and 56.1% in 2009). The other clinical symptoms were respiratory illness (6.1% in 2008 and 15.0% in 2009), acute gastroenteritis (4.9% in 2008 and 13.1% in 2009), herpangina (1.2% in 2008 and 7.5% in 2009), or hand-foot-mouth disease (1.2% in 2008 and 0.9% in 2009).
Sequence analysis of CVB 1 and CVB 5
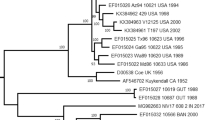
To analyze the genetic characteristics of CVB 1, the 21 CVB 1 isolates were examined by RT-PCR amplifying the VP1 region and sequencing. VP1 sequences for the CVB 1 isolates were compared with 43 foreign strains. The CVB 1 sequences segregated into four distinct clusters (A, B, C, and D) with some temporal and regional sub-clustering (Figure 3). Cluster A showed 15.7-29.2%, 19.3-32.1%, and 17.1-34.0% nucleotide divergence from cluster B, C, and D, respectively. Cluster B displayed 17.7-26.4% and 15.9-24.8% nucleotide divergence from cluster C and D, respectively. Cluster C shared 71.7-83.2% nucleotide identity with cluster D. All Korea strains (except for Kor08-CVB1-001CN) isolated from 2008 and 2009 belonged to cluster D.
The VP1 sequences of 22 CVB 5 Korean isolates obtained in 2001, 2005, and 2009 were used to construct a phylogenetic tree with 19 reference strains from the GenBank database with the same serotype. The CVB 5 strains segregated in five distinct genetic groups supported by high bootstrap values (Figure 4). Cluster A showed 18.4-21.9%, 24.9-29.6%, 15.7-21.6%, and 17.5-22.3% nucleotide divergence from cluster B, C, D, and E, respectively. Cluster B showed 22.7-27.5% 21.1-23.6%, and 15.1-16.8% nucleotide divergence from cluster C, D, and E, respectively. Cluster C showed 22.1-29.3% and 22.1-27.4% nucleotide divergence from cluster D and E, respectively. Cluster D showed 19.4-22.5% nucleotide divergence with cluster E. The 2001 Korean isolates in belonged to cluster D, whereas the Korean isolates from 2005 and 2009 belonged to cluster E. Based on the amino acid sequence comparison of the VP1 region for isolates of the CVB 5 serotype, the 2009 Korean isolates were substituted at the positions 583 (T583A). AY was substituted for GH at positions 577 and 578 in the BC loop, as shown in Figure 5.
Discussion
Various enterovirus serotypes are spreading globally by temporal and regional factors. Widely distributed throughout the year in tropical and semitropical regions, they are prominent in summer and fall, and far less detected in winter and spring months [31]. We previously reported the epidemic occurrence of enteroviruses isolated in Chungnam, Korea in 2005 and 2006 [11]. The current study extended these findings by investigating the difference of the epidemic pattern in 2008 and 2009. The data from 2008 and 2009 revealed a similar seasonal distribution in prevalence as found in 2005 and 2006. In both 2008 and 2009, enterovirus infections were most prevalent in July 42.9% and 31.9%, respectively). The highest rate of enterovirus-positive samples occurred in children < 1-year-old. However, differences with time were evident concerning the most prevalent enterovirus serotype: ECV 18 in 2005, ECV 5 in 2006, ECV 30 in 2008, and CVB 1 in 2009.
In the latest study, enteroviruses were most often isolated from patients with aseptic meningitis (80.5% in 2008 and 56.1% in 2009). Since the symptoms of enterovirus infection are quite variable, surveillance for other symptoms of enterovirus infection including aseptic meningitis could be prudent. The investigative scope for symptoms was extended in 2009; this may have reflected the marked 24% decrease in meningitis-positive samples.
PCR amplification followed by sequencing the VP1 region of the genome has been more recently used to type enteroviruses. Different VP1 sites have been targeted and demonstrated to contain serotype specific information [21, 22]. Phylogenetic analysis revealed Korean ECV 6 and ECV 30 as the prevalent enterovirus types in 2008 [32, 33]. Presently, a similar phylogenetic analysis revealed CVB 1 and 5 as the prevalent enterovirus types in Korea in 2009. Clustering or genotyping in combination with phylogenetic analysis is able to discriminate between lineages within a serotype and to identify emergent new variants or serotypes [24]. The sequences of the Korean CVB 1 isolates could be divided into four genetic clusters (A, B, C, and D) with at least 15% diversity between clusters according to PV genotypes [34]. Almost all the Korean CVB 1 isolates in 2008 and 2009 were in cluster D, with only one, "Kor08-CVB1-001CN", resident elsewhere, in cluster C. It is conceivable that "Kor08-CVB1-015CN" and "Kor08-CVB1-060CN" isolates were the origins of the 2009 Korean strains. The oldest CVB 1 isolates grouped in cluster A were viruses isolated from 1948 to the early 1980s. The viruses in clusters B and C were isolated from the 1970s to the mid-1980s and from the mid1990s to the present, respectively. The latest CVB 1 isolates clustered in group D. It seems appropriate to suggest that the grouping of these CVB 1 viruses may differ with time.
The VP1 region containing the BC loop is one of the main exposed regions of the viral capsid and has been suggested to include a serotype-specific antigenic neutralization [22, 23]. We found amino acid sequence substitutions at nine amino acid sequences (532, 562, 570, 571, 576-578, 582, 583, and 585). Especially, the only The 2009 Korean isolates displayed a T→A replacement of only amino acid 583.
In the reconstruction of the phylogenetic tree based on the VP1 nucleotide sequences of Korean isolates and the reference strains of CVB 5, all Korean isolates recovered in 2001 were segregated from the other lineage groups. In cluster E, the 2009 Korean isolates were divided to the 2005 Korean isolates because of amino acid sequence substitutions at the amino acids 578 (N→H) and 583 (T→A). Consequently, we determined the epidemiological patterns of patients with enterovirus-related diseases in Korea in 2008 and 2009. Also, the most prevalent types in 2009 (Korean CVB 1 and CVB 5) genetically characterized using amino acid sequence comparison and phylogenetic analysis.
References
Pallansch MA, Oberste MS: Molecular detection and characterization of human enteroviruses. In Cardiomyopathies and Heart Failure: Biomolecular, Infectious and Immune Mechanisms. Kluwer Academic Publishers, Boston, U.S.A: Matsumori A; 2003:245-257.
Rotbart HA: Meningitis and encephalitis. In Human Enterovirus Infections. ASM Press, Washington, D.C: Rotbart HA; 1995:271-289.
Thoelen IP, Lemey , Van der Donck I, Beuselinck K, Lindberg AM, Van Ranst M: Molecular typing and epidemiology of enteroviruses identified from an outbreak of aseptic meningitis in Belgium during the summer of 2000. J Med Virol 2003, 70: 420-429. 10.1002/jmv.10412
King AMQ, Brown F, Christian P, Hovi T, Hyypiӓ T, Knowles NJ, Lemon SM, Minor PD, Palmenberg AC, Skern T, Stanway G: Picronaviridae. In Virus taxonomy Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif: Van Regenmortel MHV, Fauquet CM, Bishop DHL, Carstens EB, Estes MK, Lemon SM, Maniloff J, Mayo MA, McGeoch DJ, Pringle CR, Wickner RB; 2000:657-678.
Mayo M, Pringle CR: Virus taxonomy. J Gen Virol 1998, 79: 649-657.
Pringle CR: Virus taxonomy at the XIth International Congress of Virology, Sydney, Australia. Arch Virol 1999, 144: 2065-2070. 10.1007/s007050050728
Stanway G, Brown F, Christian P, Hovi T, Hyypia T, King AMQ, Knowles NJ, Lemon SM, Minor PD, Pallansch MA, Palmenberg AC, Skern T: Family Picornaviridae. In Virus Taxonomy Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, London: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA; 2005:757-778.
Centers for Disease Control and Prevention: Enterovirus surveillance -- United States, 1970-2005. Morb Mortal Wkly Rep 2006, 55: 1-20.
Chomel JJ, Antona D, Thouvenot D, Lina B: Three ECHOvirus serotypes responsible for outbreak of aseptic meningitis in Rhone-Alpes region, France. Eur J Clin Microbiol Infect Dis 2003, 22: 191-193.
Jacques J, Moret H, Minette D, Leveque N, Jovenin N, Deslee G, Lebargy F, Motte J, Andreoletti L: Epidemiological, molecular, and clinical features of enterovirus respiratory infections in French children between 1999 and 2005. J Clin Microbiol 2008, 46: 206-213. 10.1128/JCM.01414-07
Baek K, Park K, Jung E, Chung E, Park J, Choi H, Baek S, Jee Y, Cheon D, Ahn G: Molecular and epidemiological characterization of enteroviruses isolated in Chungnam, Korea from 2005 to 2006J. Microbiol Biotechnol 2009,19(9):1055-1064. 10.4014/jmb.0810.584
Jee YM, Cheon DS, Choi WY, Ahn JB, Kim KS, Chung YS, Lee JW, Lee KB, Noh HS, Park KS, Lee SH, Kim SH, Cho KS, Kim ES, Jung JK, Yoon JD, Cho HW: Updates on enterovirus surveillance in Korea. Inf Chemotherapy 2004, 36: 294-303.
Caggana M, Chan P, Ramsingh A: Identification of a single amino acid residue in the capsid protein VP1 of coxsackievirus B4 that determines the virulent phenotype. J Virol 1993, 67: 4797-4803.
Dunn JJ, Chapman NM, Tracy S, Romero JR: Genomic determinants of cardiovirulence in coxsackievirus B3 clinical isolates: Localization to the 5' nontranslated region. J Virol 2000, 74: 4787-4794. 10.1128/JVI.74.10.4787-4794.2000
Knowlton KU, Jeon ES, Berkley N, Wessely R, Huber SJ: A mutation in the puff region of VP2 attenuates the myocarditic phenotype of an infectious cDNA of the Woodruff variant of coxsackievirus B3. J Virol 1996, 70: 7811-7818.
Romero JR: Reverse-transcription polymerase chain reaction detection of the enteroviruses. Arch Pathol Lab Med 1999, 123: 1161-1169.
Thoelen I, Moes E, Lemey P, Mostmans S, Wollants E, Lindberg AM, Vandamme AM, Van Ranst M: Analysis of the serotype and genotype correlation of VP1 and the 5' noncoding region in an epidemiological survey of the human enterovirus B species. J Clin Microbiol 2004, 42: 963-971. 10.1128/JCM.42.3.963-971.2004
Nix WA, Oberste MS, Pallansch MA: Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J Clin Microbiol 2006, 44: 2698-2704. 10.1128/JCM.00542-06
Oberste MS, Maher K, Kilpatrick DR, Pallansch MA: Molecular evolution of the human enteroviruses: Correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol 1999, 73: 1941-1948.
Oberste MS, Maher K, Williams AJ, Dybdahl-Sissoko N, Brown BA, Gookin MS, Peñaranda S, Mishrik N, Uddin M, Pallansch MA: Species-specific RT-PCR amplification of human enteroviruses: A tool for rapid species identification of uncharacterized enteroviruses. J Gen Virol 2006, 87: 119-128. 10.1099/vir.0.81179-0
Lee ST, Ki CS, Lee NY: Molecular characterization of enteroviruses isolated from patients with aseptic meningitis in Korea, 2005. Arch Virol 2007, 152: 963-970. 10.1007/s00705-006-0901-1
Norder H, Bjerregaard L, Magnius L, Lina B, Aymard M, Chomel JJ: Sequencing of 'untypable' enteroviruses reveals two new types, EV-77 and EV-78, within human enterovirus type B and substitutions in the BC loop of the VP1 protein for known types. J Gen Virol 2003, 84: 827-836. 10.1099/vir.0.18647-0
Stirk HJ, Thornton JM: The BC loop in poliovirus coat protein VP1: An ideal acceptor site for major insertions. Protein Eng 1994, 7: 47-56. 10.1093/protein/7.1.47
Mirand A, Archimbaud C, Henquell C, Michel Y, Chambon M, Peigue-Lafeuille H, Bailly JL: Prospective identification of HEV-B enteroviruses during the 2005 outbreak. J Med Virol 2006, 78: 1624-1634. 10.1002/jmv.20747
Zoll GJ, Melchers WJ, Kopecka H, Jambroes G, Van der Poel HJ, Galama JM: General primer-mediated polymerase chain reaction for detection of enteroviruses: Application for diagnostic routine and persistent infections. J Clin Microbiol 1992, 30: 160-165.
Oberste MS, Maher K, Flemister MR, Marchetti G, Kilpatrick DR, Pallansch MA: Comparison of classic and molecular approaches for the identification of untypeable enteroviruses. J Clin Microbiol 2000, 38: 1170-1174.
Thompson JD, Higgins DG, Gibson TJ: Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 1994, 22: 4673-4680. 10.1093/nar/22.22.4673
Martinez HM: An efficient method for finding repeats in molecular sequences. Nucleic Acids Res 1983, 11: 4629-4634. 10.1093/nar/11.13.4629
Needleman SB, Wunsch CD: A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 1970, 48: 443-453. 10.1016/0022-2836(70)90057-4
Tamura K, Dudley J, Nei M, Kumar S: MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 2007, 8: 1596-1599.
Antona D, Lévêque N, Chomel JJ, Dubrou S, Lévy-Bruhl D, Lina B: Surveillance of enteroviruses in France, 2000-2004. Eur J Clin Microbiol Infect Dis 2007, 6: 403-412.
Choi YJ, Park KS, Baek KA, Jung EH, Nam HS, Kim YB, Park JS: Molecular characterization of echovirus 30-associated outbreak of aseptic meningitis in Korea in 2008. J Microbiol Biotechnol 2010, 20: 643-649.
Jung EH, Park KS, Baek KA, Kim DU, Kang SY, Kang BH, Cheon DS: Genetic diversity of echovirus 6 strains circulating in Korea. J Bacteriol Virol 2010, 40: 191-198. 10.4167/jbv.2010.40.4.191
Rico-Hesse R, Pallansch MA, Nottay BK, Kew OM: Geographic distribution of wild poliovirus type 1 genotypes. Virology 1987, 160: 311-322. 10.1016/0042-6822(87)90001-8
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
BKA, YSG, PKS, HSY, and SJH performed molecular diagnosis and sequence analysis. LBH, YJS, RIS, KJK, and CYJ contributed to collection specimen and clinical diagnosis. CDS and PJS designed the study and critically revised the manuscript. All of the authors read and approved the final version of the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Baek, K., Yeo, S., Lee, B. et al. Epidemics of enterovirus infection in Chungnam Korea, 2008 and 2009. Virol J 8, 297 (2011). https://doi.org/10.1186/1743-422X-8-297
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1743-422X-8-297