Abstract
Background
The development of malaria vaccine has been hindered by the allele-specific responses produced by some parasite antigens’ high genetic diversity. Such antigen genetic diversity must thus be evaluated when designing a completely effective vaccine. Plasmodium falciparum P12, P38 and P41 proteins have red blood cell binding regions in the s48/45 domains and are located on merozoite surface, P41 forming a heteroduplex with P12. These three genes have been identified in Plasmodium vivax and share similar characteristics with their orthologues in Plasmodium falciparum. Plasmodium vivax pv12 and pv38 have low genetic diversity but pv41 polymorphism has not been described.
Methods
The present study was aimed at evaluating the P. vivax p41 (pv41) gene’s polymorphism. DNA sequences from Colombian clinical isolates from pv41 gene were analysed for characterising and studying the genetic diversity and the evolutionary forces that produced the variation pattern so observed.
Results
Similarly to other members of the 6-Cys family, pv41 had low genetic polymorphism. pv41 3′-end displayed the highest nucleotide diversity value; several substitutions found there were under positive selection. Negatively selected codons at inter-species level were identified in the s48/45 domains; p41 would thus seem to have functional/structural constraints due to the presence of these domains.
Conclusions
In spite of the functional constraints of Pv41 s48/45 domains, immune system pressure seems to have allowed non-synonymous substitutions to become fixed within them as an adaptation mechanism; including Pv41 s48/45 domains in a vaccine should thus be carefully evaluated due to these domains containing some allele variants.
Similar content being viewed by others
Background
Of the five malaria parasites (Plasmodium falciparum, Plasmodium vivax, Plasmodium malarie, Plasmodium ovale and Plasmodium knowlesi) affecting human beings, P. falciparum is the species causing the most severe clinical manifestations, whilst P. vivax is the species most widely distributed throughout the world, mainly affecting the Asian and American continents and causing the highest morbidity outside of Africa. In spite of efforts to date for controlling malaria, it continues to be a serious public health problem; 18.9 million cases of P. vivax occurred in 2012, children under five years old and pregnant women being the most vulnerable populations [1].
An anti-malarial vaccine represents one of the alternative control measures regarding this disease; developing a multi-antigen vaccine against the parasite’s blood stage is focused on blocking all interactions with a host cell, thereby avoiding recognition and subsequent invasion. Several antigens have been proposed as vaccine candidates [2–4]; however, as many of them have high genetic diversity [5–12], this is an obstacle regarding such proposal [13, 14] since they induce allele-specific immune responses [15]. The genetic diversity of candidate antigens must thus be evaluated [14, 16] for selecting the most frequent variants or conserved domains [13, 14].
Proteins involved in red blood cell (RBC) invasion have been characterized in merozoite surface regions known as detergent-resistant membranes (DRM) [17–19], many of these being potential vaccine candidates [4, 20, 21]. Such DRMs include a group of proteins belonging to the 6-Cys family (P12, P38, P41 and P92) which is characterised by the presence of domains containing six conserved cysteines called s48/45 [17, 22–24]. The P. falciparum P41 (Pf41) protein has two high-activity binding peptides in the s48/45 domains [17], thereby suggesting a role in RBC invasion. This protein does not have GPI-anchored domains and its presence on merozoite membrane is due to the formation of an inverted heteroduplex with Pf12 [25, 26]. The pv41 gene has recently been characterised in P. vivax (pv41) [22, 27]; this gene encodes a 385 residue-long membrane protein. Similar to its orthologue in P. falciparum, the protein has a signal peptide and two s48/45 domains but no GPI-anchor. The P. vivax P41 (Pv41) protein has been shown to be antigenic [27, 28], suggesting that it is exposed to the host immune system, probably during invasion of the host cell.
Given that Pv41 has been located on merozoite surface and that it has no membrane anchoring domains [22, 27], it could be interacting with another protein anchored to parasite surface. This protein’s similarity with its orthologue in P. falciparum suggests that Pv41 might form a complex with Pv12, a protein which has been shown to be highly conserved [29]. The present study was therefore aimed at using population genetics analysis for evaluating the pv41 gene’s genetic diversity by determining the evolutionary processes producing the locus’s variation pattern. The results showed that pv41 had low genetic diversity, the gene’s 3′-end region being the most diverse, fixing mutations by positive selection, probably as a mechanism for evading the immune system. Like other members of the 6-Cys family, this gene seemed to have functional constraints due to the presence of s48/45 domains.
Methods
Declaration of ethical considerations
This study involved using thirty P. vivax-infected samples collected between 2007 and 2010 (2007: 5 isolates, 2008: 3 isolates, 2009: 8 isolates, 2010: 14 isolates); they had been obtained from different regions of Colombia (Figure 1, South-west: Chocó, Nariño; South-east: Caquetá, Guainía, Guaviare, Meta; Midwest: Bogotá, Tolima; North-west: Atlántico, Antioquia, Córdoba). All P. vivax-infected patients who provided blood samples were notified of the study’s objective and then signed an informed consent form. All the procedures involved in taking the samples had already been approved by the Fundación Instituto de Inmunología de Colombia’s (FIDIC) ethics’ committee.
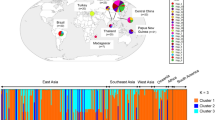
pv41 haplotype distribution in the Colombian population and origin of the reference strain sequences. Panel A shows the haplotype distribution found in pv41. Panel B shows the origin of the reference strain sequences and panel C represents haplotype frequency in the Colombian population. Haplotype 1: Sal-I, Brazil-I, haplotype 2: India, haplotype 3: Mauritania, haplotype 4: North Korea, haplotype 5: South Korea, haplotype 1–2 and 6–13: Colombian isolates.
Genotyping Plasmodium vivax samples
PCR-RFLP of the pvmsp-1 polymorphic marker was used for identifying/analysing different genotypes in the samples and infection by a single P. vivax strain, as described previously [30]. Briefly, this gene’s blocks 6, 7 and 8 were amplified with direct 5′-AAAATCGAGAGCATGATCGCCACTGAGAAG-3′ and reverse 5′-AGCTTGTACTTTCCATAGTGGTCCAG-3′ primers. The amplified fragments were digested with Alu I and Mnl I restriction enzymes.
PCR amplification of the pv41 gene
Previously reported primers were used for amplifying pv41[22]. The PCR reaction mixture contained 10 mM Tris HCl, 50 mM KCl (GeneAmp 10X PCR Buffer II (Applied Biosystems)), 1.5 mM MgCl2, 0.2 mM of each dNTP, 0.5 μM of each primer (direct 5′ ATGAAAAGGCTCCTCCTGC 3′ and reverse 5′ CTCCTGGAAGGACTTGGC 3′), 0.76 U Amplitaq Gold DNA polymerase (Applied Biosystems) and 40 ng genomic DNA at 50 μL final volume. The PCR thermal profile was as follows: one cycle at 95°C (7 min), 40 cycles at 95°C (20 sec), 60°C (30 sec), 72°C (1 min) and a final extension cycle at 72°C (10 min). The amplification products were purified using an UltraClean PCR Clean-up kit (MO BIO). The purified PCR products were bidirectionally sequenced with the amplification primers using the BigDye method with capillary electrophoresis, using the ABI-3730 XL sequencer (MACROGEN, Seoul, South Korea). Two independent PCR products were sequenced per sample to rule out errors.
Analysing genetic diversity
CLC Main workbench software v.5 (CLC bio, Cambridge, MA, USA) was used for analysing and assembling the electropherograms obtained by sequencing, giving one sequence per sample. The 30 sequences obtained from Colombian isolates were compared to and analysed regarding reference sequences obtained from several sequencing projects [31, 32] (PlasmoDB accession number: PVX_000995, GenBank accession number: AFNI01000110.1, AFNJ01000259.1, AFMK01000149.1 and AFBK01000223.1) or reported in databases (GenBank accession number: GU476495.1). These 36 sequences were then compared to Plasmodium cynomolgi (GenBank accession number: BAEJ01000104.1) and P. knowlesi orthologous sequences (PlasmoDB accession number: PKH_030970), two species which are phylogenetically close to P. vivax[33]. Gene Runner software was used for translating all the sequences for obtaining the deduced amino acid sequences; the MUSCLE algorithm was then used for aligning such sequences [34] and then edited manually. The PAL2NAL web-based tool [35] was then used for converting protein alignments into their respective nucleotide alignments.
DnaSP v.5 software [36] was used for quantifying pv41 genetic polymorphism by calculating: the number of segregant sites (Ss), the number of singleton sites (s), the number of parsimony-informative sites (Ps), the number of haplotypes (H), haplotype diversity (Hd, multiplied by (n-1)/n, according to Depaulis and Veuille [36, 37]), the Watterson estimator (θw), the average number of nucleotide differences (k) and nucleotide diversity per site (π). Data was obtained for the reference sequences plus the Colombian sequences (worldwide diversity), as well as for just the Colombian sequences (local diversity).
The Colombian parasite population sequences were used for evaluating the neutral model of molecular evolution using tests based on the frequency spectrum of nucleotide polymorphisms and haplotype distribution. Tajima’s D test [38], Fu and Li’s D* and F* tests [39], and Fay and Wu’s H test [40] were calculated for the first group of tests. Fu’s Fs test [41] and K-test and H-test [37] were calculated as part of the group of tests based on haplotype distribution. The significance of all tests was determined by coalescence simulations using DnaSP v.5 [36] and ALLELIX software (provided by Dr Sylvain Mousset). Sites having gaps were not taken into account for all tests.
The effect of natural selection was evaluated by calculating the difference between the average number of non-synonymous substitutions per non-synonymous site (dN) and the average number of synonymous substitutions per synonymous site (dS) using the modified Nei-Gojobori method [42]. Significance was determined by using Fisher’s exact tests and the Z test incorporated in MEGA v.5 software [43]. SLAC, FEL, REL [44], IFEL [45], MEME [46] and FUBAR methods [47] were used for calculating the ω (dN/dS) value for each codon in the pv41 alignment.
The McDonald-Kreitman test [48] was calculated for evaluating the effect of natural selection on p41 during the evolutionary history of P. vivax and related species (Plasmodium cynomolgi and P. knowlesi); this test compared intraspecific polymorphism with interspecific divergence using a web server [49], which takes the Jukes-Cantor distance correction regarding divergence per site [50] into account. The Nei-Gojobori modified method [42] was also used for calculating the difference between non-synonymous (KN) and synonymous (KS) divergence rates using Jukes-Cantor divergence correction [50]. Significant values were determined by using the Z test incorporate in MEGA v.5 software [43]. SLAC, FEL, REL [44], MEME [46] and FUBAR [47] methods were used for determining sites under interspecies selection using the P. vivax, P. cynomolgi and P. knowlesi sequences as data set.
ZnS[51] and ZZ [52] tests were calculated for evaluating non-random associations between polymorphisms (linkage disequilibrium or LD) and the influence of intragenic recombination on pv41. The minimum number of recombination events (Rm) [53] was also calculated and the GARD method [54] available from Datamonkey [55] was used for evaluating recombination processes.
Results
Genetic diversity in pv41
Thirty P. vivax-infected samples, obtained from different parts of Colombia (Figure 1), were genotyped using the pvmsp-1 polymorphic marker. The RFLP patterns produced from pvmsp-1 blocks 6–8 suggested the presence of different genotypes in the aforementioned samples as well as single strain infections in each sample. Taking into account that all these samples have been previously used in other studies involving genes having high polymorphism [6], in which none of the electropherograms revealed overlapping peaks during the sequencing, we can ascertain the absence of multiple infections.
The 30 genotyped isolates had a 1,152 base pair (bp) fragment corresponding to the pv41 gene. The sequences obtained from these 30 isolates (Additional file 1) were compared to and analysed together with sequences reported by several sequencing projects [31, 32]. Sequences having a different haplotype were deposited in the GenBank database (accession numbers KM212268-KM212275).
Table 1 gives the values for the estimators of genetic diversity. Seventeen segregant sites were observed in the sequences from different parts of the world, 12 of them being parsimony-informative sites and five singleton sites; 13 haplotypes were found (Figure 2). Aligning the proteins from P. vivax isolates from different geographical locations revealed substitutions in ten amino acids: N88D, E89V, A258V, Q301H, K312N, M355R, S359H, Y361F, N363D and R373G (numeration based on the Sal-I reference sequence). Ten segregant sites were found in the Colombian population (nine of them being parsimony-informative sites), giving ten haplotypes (haplotypes 1, 2, 6–13) and 0.679 ± 0.083 haplotype diversity. Haplotype 1 had 50% frequency, followed by haplotype 11 (13% frequency) and haplotype 10 (10% frequency); the remaining haplotypes had low frequency (around 3%).
The average number of nucleotide differences per pairs of sequences (k) was 3.9 when sequences from different parts of the world (worldwide diversity) were analysed and 3.1 for the Colombian population (Table 1). Low Watterson estimator (θw = 0.0038 ± 0.0009) and nucleotide diversity values (π = 0.0037 ± 0.0006) were observed when the available sequences obtained from the databases plus the Colombian ones were analysed; θw was 0.0023 ± 0.0007 and π 0.0028 ± 0.0005 for the Colombian population (Table 1). The nucleotide diversity analysis for Colombian locations showed that the Midwest was the most diverse at the pv41 locus whilst the lowest value was found in Colombia’s South-west area (Additional file 2). The gene region having the highest π value was found between nucleotides 1,064 to 1,130.
Evaluating the effect of natural selection on pv41
Tajima’s D, Fu and Li’s D* and F*, Fay and Wu’s H, Fu’s Fs and the K- and H-test neutrality tests did not give statistically significant values (Table 2); this meant that neutrality could not be ruled out. The differences between non-synonymous and synonymous (dN - dS) substitutions rates throughout the gene were evaluated for estimating the effect of natural selection in pv41, as well as in each s48/45 domain (s48/45 N-Terminal: nucleotide 76–351 and s48/45 C-Terminal: nucleotide 784–1,095); however, no significant values were found (Table 3). The sliding window (Figure 3) for the ω (dN/dS) rate gave a ω close to 1 at the 3′-end of pv41, indicating a number of non-synonymous substitutions fixed within P. vivax in this region at a higher rate than in the rest of the sequence. Tests estimating dN/dS for each site (codon) were then performed for identifying whether individual codons in pv41 were under selection; seven codons were found to be under positive selection and one codon under negative selection (Figure 3). Substitutions V269A, H312Q and G384R were exclusive for the Colombian population. The K323N, H370S amino acid changes were found in Colombian isolates and some reference sequences, whilst the N88D and E89V substitutions were present in Mauritanian and South Korean sequences, respectively.
Sliding window of omega values (ω = d N /d S and K N /K S ) for three members of the 6-Cys family expressed in merozoites. The ω values (dN/dS) for Plasmodium vivax pv12, pv38 and pv41 genes are shown in red, divergence (ω = KN/KS) between P. vivax and Plasmodium cynomolgi (Pcyn) in purple and between P. vivax and Plasmodium knowlesi (Pkno) in green. The graphical representation of each gene is given below the respective sliding window, showing the position of the segregant sites and which of them were influenced by natural selection. Intraspecies selected sites can be seen in the upper part of each gene and inter-species selected sites are indicated in the lower part. The sites were identified by the Datamonkey server. The schemes for pv12 and pv38 have been modified from Forero-Rodríguez et al. [29].
The McDonald-Kreitman test was calculated for evaluating how selection had acted throughout p41’s evolutionary history; it revealed significant values, thereby showing that polymorphism was greater than divergence (p < 0.05) (Table 4). A sliding window for ω divergence (KN/KS, non-synonymous divergence/synonymous divergence), obtained by comparing the P. vivax sequences to sequences from phylogenetically close species (P. cynomolgi and P. knowlesi), gave values less than 1 in the s48/45 domains, as well as in some areas between these domains, thereby indicating that KS tended to be greater than KN. Significant negative values (p < 0.001) were found when estimating the difference between non-synonymous and synonymous divergence (KN - KS) (Table 5). The codon-based selection tests found 13 positively selected codons and 77 negatively selected codons at inter-species level (Figure 3).
Linkage disequilibrium (LD) and recombination
The ZnS, ZZ and RM tests were calculated for determining possible associations between polymorphism and/or the presence of recombination in pv41 (Table 2). The ZnS test gave 0.3627, this being statistically significant (p < 0.05). Lineal regression between LD and nucleotide distance gave a slight reduction in LD as nucleotide distance increased, suggesting recombination events. This was confirmed when the ZZ test was calculated, giving 0.2073 (p < 0.05); two minimum recombination sites were found (Table 2). The GARD method (available from the Datamonkey web server) gave a recombination breakpoint in position 936 (number based on Sal-I sequence) confirming than intragenic recombination was involved in generating new haplotypes in pv41.
Discussion
Merozoite-expressed members of the 6-Cys family in P. falciparum (Pf12, Pf38 and Pf41) have high RBC binding activity peptides [17], indicating that these play a role during recognition of a host cell. Previous studies have shown that members of this family are antigenic [23, 24, 27, 28] and highly conserved (p12 and p38) in both P. falciparum and P. vivax[26, 29, 56, 57]. This means that they are promising candidates for inclusion in an anti-malarial vaccine, avoiding allele-specific immune responses. The pv41 gene has been shown to be highly conserved when compared to other genes encoding antigens in P. vivax (e.g., pvmsp-7[6], pvmsp-5[7, 12], pvmsp-3[9, 10], pvmsp-1[5, 8]).
The pv41 nucleotide diversity was low in the Colombian population; however, π values and haplotype number were dissimilar for each Colombian locality, suggesting different evolutionary histories possibly due to a structured population. However, this pattern could have been due to few samples having been collected from some locations. The use of neutral markers could lead to confirming whether Colombia has a structured population.
pv41 nucleotide diversity was higher than that reported for pv12, but similar to that found in pv38[29]; however, fewer haplotypes were found in pv41 compared to pv38 (14 haplotypes have been reported for it in the Colombian population) [29]. Since the Pv41 protein has no membrane-anchoring domains, it could be interacting with proteins anchored to the merozoite surface. It has been shown that Pf12 and Pf41 proteins form an inverted heteroduplex on parasite membrane [25, 26]. Due to these proteins’ similarity, it is probable that Pv12 and Pv41 may also interact in P. vivax. This could explain the high degree of conservation found in Pv12 (π = 0.0004 ± 0.0001 [29]). If Pv41 forms a protein complex with Pv12, the latter could be masked whilst Pv41 would be more exposed to a host’s immune system, greater diversity thus being found in Pv41 (π = 0.0037 ± 0.0006) regarding Pv12 (π = 0.0004 ± 0.0001). Since such complex formation would be anti-parallel, the region most exposed to Pv41 would be the C-terminal in which high fixation of non-synonymous substitutions was observed (Figure 3).
No significant values were found in the neutrality tests based on the polymorphism frequency spectrum or the haplotype-based tests (Table 2), meaning that the hypothesis regarding neutrality could not be ruled out. Such hypothesis stated that pv41 haplotypes could be fixed in different populations thereby producing a population structure in this locus and new pv41 haplotypes might thus appear if new parasites populations are evaluated.
No significant values were found when the effect of natural selection was evaluated by means of the difference between non-synonymous and synonymous substitutions (dN - dS) in either the whole gene or in each s48/45 domain (Table 3). However, the pv41 sliding window gave a peak close to 1 at the 3′-end of the gene (Figure 3); several non-synonymous mutations would thus seem to be fixed in this region. The codon-based selection tests showed that seven out of the ten codons having mutations producing a change in the protein were positively selected (Figure 3). Three of these seven codons (V89E, H359S and G373R) produced radical substitutions (changing amino acid physical/chemical properties). The R355M substitution also produced a radical change but selection signals were not identified in this site. Such positively selected codons were predominantly found towards the gene’s 3′-end (encoding the protein’s C-terminal region) and could have been fixed to enable evading the immune system since this region would be more exposed due to the possible antiparallel formation of a Pv12/Pv41complex. Substitutions in codons 258, 301 and 312 located in the s48/45 domain could become deleterious due to them being able to alter the domain’s structure; however, they had positive selection signals. Such substitutions were conservative and maintained the amino acids’ physical-chemical characteristics, thereby enabling evasion of the immune system and maintaining the domain’s structural conformation. Interspecies ω values were higher than 1 in some regions of p41, mainly outside s48/45 domains. Thirteen codons were positively selected at interspecies level; amino acid fixation would allow immune evasion of the respective host. Alternatively, positive sites found in s48/45 domains (which are involved in red blood cell invasion [17]) would be a P41 adaptation to the host receptor molecule.
The ZnS test had significant values, indicating LD. The linked positions were found in the 3′-end of the gene. The mutations found there led to changes in protein sequence H359S, Y361F and D363N. The first substitution (H359S) produced a radical amino acid change, which was fixed by positive selection whilst the other two changes were conservative without selection signals. Since amino acid H359S was fixed by positive selection, this led to Y361F and D363N becoming fixed due to the short physical distance between them.
Genetic diversity in pv41 was produced by point mutations (Figure 2); however, the recombination could also have been responsible for the genetic polymorphism found in this gene. The lineal regression between LD and nucleotide distance had a slight reduction in LD as nucleotide distance increased; this may have been a consequence of recombination processes. The ZZ test gave significant values, suggesting that recombination took place in this gene. Two minimum recombination sites were found and the GARD method (available from the Datamonkey web server) identified a recombination breakpoint in position 936, meaning that recombination produced new haplotypes in pv41.
The McDonald-Kreitman (MK) and omega divergence tests (ω = KN/KS) were calculated for inferring natural selection signals which might have influence the evolutionary history of p41. The latter was calculated for the gene’s complete length and for each s48/45 domain. Significant values were found in the MK test throughout the whole gene (Table 5), polymorphism being greater than divergence; this could have resulted from weak negative selection or balancing positive selection. The latter is responsible for keeping allele variants (haplotypes) at intermediate frequencies as a mechanism for evading host immune responses; however, a major haplotype was found in the Colombian population whilst the rest occurred at low frequency. Due to the population structure reported in America [58], haplotype segregation could have led to different frequencies or new haplotypes could have diversified within American (or Colombian) subpopulations, meaning that if just one population is analysed, then balancing positive selection signals will not always be detected with population methods (Tajima, Fu and Li, Fay and Wu, Fu and K-test, and H-test). Alternatively, if balancing selection has resulted from frequency dependent selection, it would be expected that a haplotype would be presented as a major allele during a determined period of time and then become replaced by another less frequent one as an evasion mechanism. These haplotypes’ frequency must therefore be evaluated during different intervals of time in several populations involving larger sampling.
On the other hand, the ω (KN/KS) rate sliding window showed that most values obtained throughout the gene were lower than 1, indicating high synonymous substitution fixation following P. vivax/P. cynomolgi/P. knowlesi divergence. The same pattern was observed in pv12 and pv38 (Figure 3 and [29]). The difference between non-synonymous and synonymous (KN - KS) divergence was estimated, giving significant negative values (p < 0.001) in pv41 as well as in the s48/45 domains of this gene (Table 4). A large amount of negatively selected codons were identified which were preferentially located in the s48/45 domains (Figure 3). These results suggested that p41 had diverged due to negative selection; such pattern was similar to that previously reported for other members of the P. vivax 6-Cys family [29, 56]. pv12 and pv38, like pv41, had various codons under negative selection at interspecies level which were preferentially located in the s48/45 domains (Figure 3). Such accumulation of interspecies synonymous substitutions suggested that evolution had tried to maintain domain structure in the different members of the 6-Cys family by eliminating all deleterious mutations due to the functional importance which these domains seem to have [17, 59].
Conclusions
6-Cys family members seem to play a role during host cell recognition [17, 59]. Due to the high degree of P12, P38 [29] and P41 protein conservation (at both intraspecies and interspecies level) given by the fixation of a large amount of synonymous substitutions, these three proteins may have evolved under strong functional constraints, possibly due to the presence of s48/45 domains which seem to have served as ligands for recognising the host cell [17, 59, 60]. Consequently, s48/45 domains should remain conserved as the resulting mutations could be deleterious; their evolution would thus have been slower regarding other functionally less important ones. Pv12, Pv38 and Pv41 thus warrant consideration as valuable candidates for developing a vaccine. However, a functional constraint does not imply that these regions may not vary. Pv41 s48/45 domains have been seen to have changes in their protein sequence, which seem to have been positively selected. Such changes conserve physical-chemical properties and thus structure/function may not become compromised, but could enable evasion of the immune response. Including Pv41 in a vaccine should thus be carefully evaluated due to the presence of variants in these regions.
This is also another aspect that must be taken into account when developing vaccines. It has been proposed that a completely effective vaccine requires the inclusion of both functional and conserved regions; however, vaccination could thus produce new selective pressure in these regions and parasites could fix mutations as an adaptation mechanism (in spite of their functional importance) and the appearance of new variants might thus reduce vaccine’s efficacy.
References
WHO: World Malaria Report 2013. 2013, Geneva: World Health Organization,http://www.who.int/malaria/publications/world_malaria_report_2013/wmr2013_no_profiles.pdf,
Carvalho LJ, Daniel-Ribeiro CT, Goto H: Malaria vaccine: candidate antigens, mechanisms, constraints and prospects. Scand J Immunol. 2002, 56: 327-343. 10.1046/j.1365-3083.2002.01160.x.
Jones TR, Hoffman SL: Malaria vaccine development. Clin Microbiol Rev. 1994, 7: 303-310.
Patarroyo MA, Calderon D, Moreno-Perez DA: Vaccines against Plasmodium vivax: a research challenge. Expert Rev Vaccines. 2012, 11: 1249-1260. 10.1586/erv.12.91.
Figtree M, Pasay CJ, Slade R, Cheng Q, Cloonan N, Walker J, Saul A: Plasmodium vivax synonymous substitution frequencies, evolution and population structure deduced from diversity in AMA 1 and MSP 1 genes. Mol Biochem Parasitol. 2000, 108: 53-66. 10.1016/S0166-6851(00)00204-8.
Garzon-Ospina D, Lopez C, Forero-Rodriguez J, Patarroyo MA: Genetic diversity and selection in three Plasmodium vivax merozoite surface protein 7 (Pvmsp-7) genes in a Colombian population. PLoS One. 2012, 7: e45962-10.1371/journal.pone.0045962.
Gomez A, Suarez CF, Martinez P, Saravia C, Patarroyo MA: High polymorphism in Plasmodium vivax merozoite surface protein-5 (MSP5). Parasitology. 2006, 133: 661-672. 10.1017/S0031182006001168.
Kang JM, Ju HL, Kang YM, Lee DH, Moon SU, Sohn WM, Park JW, Kim TS, Na BK: Genetic polymorphism and natural selection in the C-terminal 42 kDa region of merozoite surface protein-1 among Plasmodium vivax Korean isolates. Malar J. 2012, 11: 206-10.1186/1475-2875-11-206.
Mascorro CN, Zhao K, Khuntirat B, Sattabongkot J, Yan G, Escalante AA, Cui L: Molecular evolution and intragenic recombination of the merozoite surface protein MSP-3alpha from the malaria parasite Plasmodium vivax in Thailand. Parasitology. 2005, 131: 25-35. 10.1017/S0031182005007547.
Ord R, Polley S, Tami A, Sutherland CJ: High sequence diversity and evidence of balancing selection in the Pvmsp3alpha gene of Plasmodium vivax in the Venezuelan Amazon. Mol Biochem Parasitol. 2005, 144: 86-93. 10.1016/j.molbiopara.2005.08.005.
Putaporntip C, Jongwutiwes S, Seethamchai S, Kanbara H, Tanabe K: Intragenic recombination in the 3′ portion of the merozoite surface protein 1 gene of Plasmodium vivax. Mol Biochem Parasitol. 2000, 109: 111-119. 10.1016/S0166-6851(00)00238-3.
Putaporntip C, Udomsangpetch R, Pattanawong U, Cui L, Jongwutiwes S: Genetic diversity of the Plasmodium vivax merozoite surface protein-5 locus from diverse geographic origins. Gene. 2010, 456: 24-35. 10.1016/j.gene.2010.02.007.
Richie TL, Saul A: Progress and challenges for malaria vaccines. Nature. 2002, 415: 694-701. 10.1038/415694a.
Takala SL, Plowe CV: Genetic diversity and malaria vaccine design, testing and efficacy: preventing and overcoming ‘vaccine resistant malaria’. Parasite Immunol. 2009, 31: 560-573. 10.1111/j.1365-3024.2009.01138.x.
Polley SD, Tetteh KK, Lloyd JM, Akpogheneta OJ, Greenwood BM, Bojang KA, Conway DJ: Plasmodium falciparum merozoite surface protein 3 is a target of allele-specific immunity and alleles are maintained by natural selection. J Infect Dis. 2007, 195: 279-287. 10.1086/509806.
Arnott A, Barry AE, Reeder JC: Understanding the population genetics of Plasmodium vivax is essential for malaria control and elimination. Malar J. 2012, 11: 14-10.1186/1475-2875-11-14.
Garcia J, Curtidor H, Pinzon CG, Vanegas M, Moreno A, Patarroyo ME: Identification of conserved erythrocyte binding regions in members of the Plasmodium falciparum Cys6 lipid raft-associated protein family. Vaccine. 2009, 27: 3953-3962. 10.1016/j.vaccine.2009.04.039.
Nagao E, Seydel KB, Dvorak JA: Detergent-resistant erythrocyte membrane rafts are modified by a Plasmodium falciparum infection. Exp Parasitol. 2002, 102: 57-59. 10.1016/S0014-4894(02)00143-1.
Sanders PR, Gilson PR, Cantin GT, Greenbaum DC, Nebl T, Carucci DJ, McConville MJ, Schofield L, Hodder AN, Yates JR, Crabb BS: Distinct protein classes including novel merozoite surface antigens in Raft-like membranes of Plasmodium falciparum. J Biol Chem. 2005, 280: 40169-40176. 10.1074/jbc.M509631200.
Barrero CA, Delgado G, Sierra AY, Silva Y, Parra-Lopez C, Patarroyo MA: Gamma interferon levels and antibody production induced by two PvMSP-1 recombinant polypeptides are associated with protective immunity against P. vivax in Aotus monkeys. Vaccine. 2005, 23: 4048-4053. 10.1016/j.vaccine.2005.02.012.
Richards JS, Beeson JG: The future for blood-stage vaccines against malaria. Immunol Cell Biol. 2009, 87: 377-390. 10.1038/icb.2009.27.
Angel DI, Mongui A, Ardila J, Vanegas M, Patarroyo MA: The Plasmodium vivax Pv41 surface protein: identification and characterization. Biochem Biophys Res Commun. 2008, 377: 1113-1117. 10.1016/j.bbrc.2008.10.129.
Mongui A, Angel DI, Guzman C, Vanegas M, Patarroyo MA: Characterisation of the Plasmodium vivax Pv38 antigen. Biochem Biophys Res Commun. 2008, 376: 326-330. 10.1016/j.bbrc.2008.08.163.
Moreno-Perez DA, Areiza-Rojas R, Florez-Buitrago X, Silva Y, Patarroyo ME, Patarroyo MA: The GPI-anchored 6-Cys protein Pv12 is present in detergent-resistant microdomains of Plasmodium vivax blood stage schizonts. Protist. 2013, 164: 37-48. 10.1016/j.protis.2012.03.001.
Taechalertpaisarn T, Crosnier C, Bartholdson SJ, Hodder AN, Thompson J, Bustamante LY, Wilson DW, Sanders PR, Wright GJ, Rayner JC, Cowman AF, Gilson PR, Crabb BS: Biochemical and functional analysis of two Plasmodium falciparum blood-stage 6-cys proteins: P12 and P41. PLoS One. 2012, 7: e41937-10.1371/journal.pone.0041937.
Tonkin ML, Arredondo SA, Loveless BC, Serpa JJ, Makepeace KA, Sundar N, Petrotchenko EV, Miller LH, Grigg ME, Boulanger MJ: Structural and biochemical characterization of Plasmodium falciparum 12 (Pf12) reveals a unique interdomain organization and the potential for an antiparallel arrangement with Pf41. J Biol Chem. 2013, 288: 12805-12817. 10.1074/jbc.M113.455667.
Cheng Y, Lu F, Tsuboi T, Han ET: Characterization of a novel merozoite surface protein of Plasmodium vivax, Pv41. Acta Trop. 2013, 126: 222-228. 10.1016/j.actatropica.2013.03.002.
Chen JH, Jung JW, Wang Y, Ha KS, Lu F, Lim CS, Takeo S, Tsuboi T, Han ET: Immunoproteomics profiling of blood stage Plasmodium vivax infection by high-throughput screening assays. J Proteome Res. 2010, 9: 6479-6489. 10.1021/pr100705g.
Forero-Rodriguez J, Garzon-Ospina D, Patarroyo MA: Low genetic diversity and functional constraint in loci encoding Plasmodium vivax P12 and P38 proteins in the Colombian population. Malar J. 2014, 13: 58-10.1186/1475-2875-13-58.
Imwong M, Pukrittayakamee S, Gruner AC, Renia L, Letourneur F, Looareesuwan S, White NJ, Snounou G: Practical PCR genotyping protocols for Plasmodium vivax using Pvcs and Pvmsp1. Malar J. 2005, 4: 20-10.1186/1475-2875-4-20.
Neafsey DE, Galinsky K, Jiang RH, Young L, Sykes SM, Saif S, Gujja S, Goldberg JM, Young S, Zeng Q, Chapman SB, Dash AP, Anvikar AR, Sutton PL, Birren BW, Escalante AA, Barnwell JW, Carlton JM: The malaria parasite Plasmodium vivax exhibits greater genetic diversity than Plasmodium falciparum. Nat Genet. 2012, 44: 1046-1050. 10.1038/ng.2373.
Bozdech Z, Mok S, Hu G, Imwong M, Jaidee A, Russell B, Ginsburg H, Nosten F, Day NP, White NJ, Carlton JM, Preiser PR: The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc Natl Acad Sci U S A. 2008, 105: 16290-16295. 10.1073/pnas.0807404105.
Pacheco MA, Battistuzzi FU, Junge RE, Cornejo OE, Williams CV, Landau I, Rabetafika L, Snounou G, Jones-Engel L, Escalante AA: Timing the origin of human malarias: the lemur puzzle. BMC Evol Biol. 2011, 11: 299-10.1186/1471-2148-11-299.
Edgar RC: MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32: 1792-1797. 10.1093/nar/gkh340.
Suyama M, Torrents D, Bork P: PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006, 34: W609-W612. 10.1093/nar/gkl315.
Librado P, Rozas J: DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009, 25: 1451-1452. 10.1093/bioinformatics/btp187.
Depaulis F, Veuille M: Neutrality tests based on the distribution of haplotypes under an infinite-site model. Mol Biol Evol. 1998, 15: 1788-1790. 10.1093/oxfordjournals.molbev.a025905.
Tajima F: Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989, 123: 585-595.
Fu YX, Li WH: Statistical tests of neutrality of mutations. Genetics. 1993, 133: 693-709.
Fay JC, Wu CI: Hitchhiking under positive Darwinian selection. Genetics. 2000, 155: 1405-1413.
Fu YX: Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997, 147: 915-925.
Zhang J, Rosenberg HF, Nei M: Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc Natl Acad Sci U S A. 1998, 95: 3708-3713. 10.1073/pnas.95.7.3708.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S: MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011, 28: 2731-2739. 10.1093/molbev/msr121.
Kosakovsky Pond SL, Frost SD: Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005, 22: 1208-1222. 10.1093/molbev/msi105.
Pond SL, Frost SD, Grossman Z, Gravenor MB, Richman DD, Brown AJ: Adaptation to different human populations by HIV-1 revealed by codon-based analyses. PLoS Comput Biol. 2006, 2: e62-10.1371/journal.pcbi.0020062.
Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL: Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012, 8: e1002764-10.1371/journal.pgen.1002764.
Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Kdosakovsky Pond SL, Scheffler K: FUBAR: a fast, unconstrained bayesian approximation for inferring selection. Mol Biol Evol. 2013, 30: 1196-1205. 10.1093/molbev/mst030.
McDonald JH, Kreitman M: Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991, 351: 652-654. 10.1038/351652a0.
Egea R, Casillas S, Barbadilla A: Standard and generalized McDonald-Kreitman test: a website to detect selection by comparing different classes of DNA sites. Nucleic Acids Res. 2008, 36: W157-W162. 10.1093/nar/gkn337.
Jukes THCRC: Evolution of Protein Molecules. Mammalian Protein Metabolism. Edited by: Munro HN. 1969, New York: Academic Press
Kelly JK: A test of neutrality based on interlocus associations. Genetics. 1997, 146: 1197-1206.
Rozas J, Gullaud M, Blandin G, Aguade M: DNA variation at the rp49 gene region of Drosophila simulans: evolutionary inferences from an unusual haplotype structure. Genetics. 2001, 158: 1147-1155.
Hudson RR, Kaplan NL: Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985, 111: 147-164.
Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD: Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol. 2006, 23: 1891-1901. 10.1093/molbev/msl051.
Delport W, Poon AF, Frost SD, Kosakovsky Pond SL: Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics. 2010, 26: 2455-2457. 10.1093/bioinformatics/btq429.
Doi M, Tanabe K, Tachibana S, Hamai M, Tachibana M, Mita T, Yagi M, Zeyrek FY, Ferreira MU, Ohmae H, Kaneko A, Randrianarivelojosia M, Sattabongkot J, Cao YM, Horii T, Torii M, Tsuboi T: Worldwide sequence conservation of transmission-blocking vaccine candidate Pvs230 in Plasmodium vivax. Vaccine. 2011, 29: 4308-4315. 10.1016/j.vaccine.2011.04.028.
Reeder JC, Wapling J, Mueller I, Siba PM, Barry AE: Population genetic analysis of the Plasmodium falciparum 6-cys protein Pf38 in Papua New Guinea reveals domain-specific balancing selection. Malar J. 2011, 10: 126-10.1186/1475-2875-10-126.
Taylor JE, Pacheco MA, Bacon DJ, Beg MA, Machado RL, Fairhurst RM, Herrera S, Kim JY, Menard D, Povoa MM, Villegas L, Mulyanto , Snounou G, Cui L, Zeyrek FY, Escalante AA: The evolutionary history of Plasmodium vivax as inferred from mitochondrial genomes: parasite genetic diversity in the Americas. Mol Biol Evol. 2013, 30: 2050-2064. 10.1093/molbev/mst104.
Cowman AF, Crabb BS: Invasion of red blood cells by malaria parasites. Cell. 2006, 124: 755-766. 10.1016/j.cell.2006.02.006.
Arredondo SA, Cai M, Takayama Y, MacDonald NJ, Anderson DE, Aravind L, Clore GM, Miller LH: Structure of the Plasmodium 6-cysteine s48/45 domain. Proc Natl Acad Sci U S A. 2012, 109: 6692-6697. 10.1073/pnas.1204363109.
Acknowledgements
We would like to thank Dr Sylvain Mousset for providing the Allelix software for our analysis, Professor María Isabel Chacón for her comments and suggestions and Jason Garry for translating this manuscript. This work was financed by the “Departamento Administrativo de Ciencia, Tecnología e Innovación (Colciencias)” through contracts RC #0309–2013 and 709–2013. JF-R received funding from COLCIENCIAS via cooperation agreement #0719–2013.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JF-R devised the study, participated in designing it, performed the experiments, made the population genetics analysis and wrote the manuscript. DG-O devised and designed the study, helped perform the experiments, carried out the population genetics analysis and wrote the manuscript. MAP coordinated the study, and helped to write the manuscript. All the authors have read and approved the final manuscript.
Electronic supplementary material
12936_2014_3548_MOESM2_ESM.pdf
Additional file 2: Nucleotide diversity (π) values for subpopulations within Colombia. n: number of isolates, Ss: number of segregant sites, S: number of singleton sites, Ps: number of parsimony-informative sites, H: number of haplotypes, k: average number of nucleotide differences by sequence pairs, π: nucleotide diversity per site. (PDF 113 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Forero-Rodríguez, J., Garzón-Ospina, D. & Patarroyo, M.A. Low genetic diversity in the locus encoding the Plasmodium vivax P41 protein in Colombia’s parasite population. Malar J 13, 388 (2014). https://doi.org/10.1186/1475-2875-13-388
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2875-13-388