Abstract
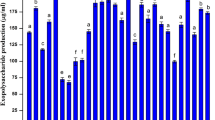
Saline environments are extreme habitats with a high diversity of microorganisms source of a myriad of biomolecules. These microorganisms are assigned as extremophiles recognized to be producers of new natural compounds, which can be synthesized by helping to survive under harshness and extreme conditions. In Brazil, in the saline and semi-arid region of Areia Branca (Caatinga biome), halotolerant bacteria (able to growth at high NaCl concentrations) were isolated from rhizosphere of native plants Blutaparon portulacoides and Spergularia sp. and their biopolymer production was studied. A total of 25 bacterial isolates were identified at genus level based on 16S rRNA gene sequence analysis. Isolates were mainly Gram-positive bacteria from Bacillaceae, Staphylococcaceae, Microbacteriaceae, and Bacillales XII incertae sedis families, affiliates to Bacillus, Staphylococcus, Curtobacterium, and Exiguobacterium genera, respectively. One of the Gram-negative isolates was identified as member of the Pseudomonadaceae family, genus Pseudomonas. All the identified strains were halotolerant bacteria with optimum growth at 0.6–2.0 M salt concentrations. Assays for biopolymer production showed that the halotolerant strains are a rich source of compounds as polyhydroxyalkanoates (PHA), biodegradable biopolymer, such as poly(3-hydroxybutyrate) (PHB) produced from low-cost substrates, and exopolysaccharides (EPS), such as hyaluronic acid (HA), metabolite of great interest to the cosmetic and pharmaceutical industry. Also, eight bacterial EPS extracts showed immunostimulatory activity, promising results that can be used in biomedical applications. Overall, our findings demonstrate that these biomolecules can be produced in culture medium with 0.6–2.0 M NaCl concentrations, relevant feature to avoid costly production processes. This is the first report of biopolymer-producing bacteria from a saline region of Caatinga biome that showed important biological activities.






Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
References
Corral P, Amoozegar MA, Ventosa A (2020) Halophiles and their biomolecules: recent advances and future applications in biomedicine. Mar Drugs 18. https://doi.org/10.3390/md18010033
Kavamura VN, Taketani RG, Ferreira C, de Melo IS, Mendes R (2018) The role of species turnover in structuring bacterial communities in a local scale in the cactus rhizosphere. Plant Soil 425:101–112. https://doi.org/10.1007/s11104-018-3570-4
Ricardo SDF, Coe HHG, Dias RR, de Sousa LOF, Gomes E (2018) Reference collection of plant phytoliths from the Caatinga biome, Northeast Brazil. Flora Morphol Distrib Funct Ecol Plants 249:1–8. https://doi.org/10.1016/j.flora.2018.09.003
Taketani RG, Lançoni MD, Kavamura VN, Durrer A, Andreote FD, Melo IS (2017) Dry season constrains bacterial phylogenetic diversity in a semi-arid rhizosphere system. Microb Ecol 73:153–161. https://doi.org/10.1007/s00248-016-0835-4
Hamedi J, Mohammadipanah F, Ventosa A (2013) Systematic and biotechnological aspects of halophilic and halotolerant actinomycetes. Extremophiles 17:1–13. https://doi.org/10.1007/s00792-012-0493-5
Gunde-Cimerman N, Plemenitaš A, Oren A (2018) Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol Rev 42:353–375. https://doi.org/10.1093/femsre/fuy009
Corinaldesi C, Barone G, Marcellini F et al (2017) Marine microbial-derived molecules and their potential use in cosmeceutical and cosmetic products. 1–21. https://doi.org/10.3390/md15040118
Liu C, Baffoe DK, Zhan Y, Zhang M, Li Y, Zhang G (2019) Halophile, an essential platform for bioproduction. J Microbiol Methods 166:1–8. https://doi.org/10.1016/j.mimet.2019.105704
Schmid MT, Song H, Raschbauer M, Emerstorfer F, Omann M, Stelzer F, Neureiter M (2019) Utilization of desugarized sugar beet molasses for the production of poly(3-hydroxybutyrate) by halophilic Bacillus megaterium uyuni S29. Process Biochem 86:9–15. https://doi.org/10.1016/j.procbio.2019.08.001
Boujida N, Palau M, Charfi S, el Moussaoui N, Manresa A, Miñana-Galbis D, Skali Senhaji N, Abrini J (2018) Isolation and characterization of halophilic bacteria producing exopolymers with emulsifying and antioxidant activities. Biocatal Agric Biotechnol 16:631–637. https://doi.org/10.1016/j.bcab.2018.10.015
Radchenkova N, Boyadzhieva I, Atanasova N, Poli A, Finore I, di Donato P, Nicolaus B, Panchev I, Kuncheva M, Kambourova M (2018) Extracellular polymer substance synthesized by a halophilic bacterium Chromohalobacter canadensis 28. Appl Microbiol Biotechnol 102:4937–4949. https://doi.org/10.1007/s00253-018-8901-0
Powell JT, Chatziefthimiou AD, Banack SA, Cox PA, Metcalf JS (2015) Desert crust microorganisms, their environment, and human health. J Arid Environ 112:127–133. https://doi.org/10.1016/j.jaridenv.2013.11.004
Yoon S-H, Ha S-M, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A 101:11030–11035. https://doi.org/10.1073/pnas.0404206101
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376. https://doi.org/10.1007/BF01734359
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10. https://doi.org/10.1093/oxfordjournals.molbev.a040023
Ramsay BA, Lomaliza K, Chavarie C et al (1990) Production of poly-(beta-hydroxybutyric-co-beta-hydroxyvaleric) acids. Appl Environ Microbiol 56:2093–2098. https://doi.org/10.1016/S0922-338X(97)83009-7
Schlegel HG, Lafferty R, Krauss I (1970) The isolation of mutants not accumulating poly-beta-hydroxybutyric acid. Arch Mikrobiol 71:283–294. https://doi.org/10.1007/BF00410161
Riis V, Mai W (1988) Gas chromatographic determination of poly-β-hydroxybutyric acid in microbial biomass after hydrochloric acid propanolysis. J Chromatogr A 445:285–289. https://doi.org/10.1016/S0021-9673(01)84535-0
Mendonça TT, Tavares RR, Cespedes LG, Sánchez-Rodriguez RJ, Schripsema J, Taciro MK, Gomez JGC, Silva LF (2017) Combining molecular and bioprocess techniques to produce poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) with controlled monomer composition by Burkholderia sacchari. Int J Biol Macromol 98:654–663. https://doi.org/10.1016/j.ijbiomac.2017.02.013
Paulo EM, Vasconcelos MP, Oliveira IS, Affe HMJ, Nascimento R, Melo IS, Roque MRA, Assis SA (2012) An alternative method for screening lactic acid bacteria for the production of exopolysaccharides with rapid confirmation. Food Sci Technol 32:710–714. https://doi.org/10.1590/S0101-20612012005000094
Kavamura VN, Santos SN, da Silva JL et al (2013) Screening of Brazilian cacti rhizobacteria for plant growth promotion under drought. Microbiol Res 168:183–191. https://doi.org/10.1016/j.micres.2012.12.002
Liang TW, Wang SL (2015) Recent advances in exopolysaccharides from Paenibacillus spp.: production, isolation, structure, and bioactivities. Mar Drugs 13:1847–1863. https://doi.org/10.3390/md13041847
Green LC, Tannenbaum SR, Goldman P (1981) Nitrate synthesis in the germfree and conventional rat. Science 212:56–58. https://doi.org/10.1126/science.6451927
Yu H, Tyo K, Alper H, Klein-Marcuschamer D, Stephanopoulos G (2008) A high-throughput screen for hyaluronic acid accumulation in recombinant Escherichia coli transformed by libraries of engineered sigma factors. Biotechnol Bioeng 101:788–796. https://doi.org/10.1002/bit.21947
Zahir ZA, Nadeem SM, Khan MY, Binyamin R (2019) Role of halotolerant microbes in plant growth promotion under salt stress conditions. In: Kumar M, Etesami H, Kumar V (eds) Saline soil-based agriculture by halotolerant microorganisms. Springer, Singapore, pp 209–253
Mukhtar S, Mehnaz S, Mirza MS, Malik KA (2019) Isolation and characterization of bacteria associated with the rhizosphere of halophytes (Salsola stocksii and Atriplex amnicola) for production of hydrolytic enzymes. Braz J Microbiol 50:85–97. https://doi.org/10.1007/s42770-019-00044-y
Silva-Lacerda GR, Santana RCF, Vicalvi-Costa MCV, Solidônio EG, Sena KXFR, Lima GMS, Araújo JM (2016) Antimicrobial potential of actinobacteria isolated from the rhizosphere of the Caatinga biome plant Caesalpinia pyramidalis Tul. Genet Mol Res 15:1–12. https://doi.org/10.4238/gmr.15017488
Abbas R, Rasul S, Aslam K, Baber M, Shahid M, Mubeen F, Naqqash T (2019) Halotolerant PGPR: a hope for cultivation of saline soils. J King Saud Univ - Sci 31:1195–1201. https://doi.org/10.1016/j.jksus.2019.02.019
López G, Diaz-Cárdenas C, Shapiro N, Woyke T, Kyrpides NC, David Alzate J, González LN, Restrepo S, Baena S (2017) Draft genome sequence of Pseudomonas extremaustralis strain USBA-GBX 515 isolated from superparamo soil samples in Colombian Andes. Stand Genomic Sci 12:1–12. https://doi.org/10.1186/s40793-017-0292-9
Cheng Y, Xiao X, Li X, Song D, Lu Z, Wang F, Wang Y (2017) Characterization, antioxidant property and cytoprotection of exopolysaccharide-capped elemental selenium particles synthesized by Bacillus paralicheniformis SR14. Carbohydr Polym 178:18–26. https://doi.org/10.1016/j.carbpol.2017.08.124
Mohandas SP, Balan L, Jayanath G, Anoop BS, Philip R, Cubelio SS, Bright Singh IS (2018) Biosynthesis and characterization of polyhydroxyalkanoate from marine Bacillus cereus MCCB 281 utilizing glycerol as carbon source. Int J Biol Macromol 119:380–392. https://doi.org/10.1016/j.ijbiomac.2018.07.044
Bhatnagar A, Bhatnagar M (2005) Microbial diversity in desert ecosystems. Curr Sci 89:91–100. https://doi.org/10.1016/S1369-5274(02)00324-7
Kuhlman KR, Allenbach LB, Ball CL, Fusco WG, la Duc MT, Kuhlman GM, Anderson RC, Stuecker T, Erickson IK, Benardini J, Crawford RL (2005) Enumeration, isolation, and characterization of ultraviolet (UV-C) resistant bacteria from rock varnish in the Whipple Mountains, California. Icarus 174:585–595. https://doi.org/10.1016/j.icarus.2004.11.022
Cardinale M, Ratering S, Suarez C, Zapata Montoya AM, Geissler-Plaum R, Schnell S (2015) Paradox of plant growth promotion potential of rhizobacteria and their actual promotion effect on growth of barley (Hordeum vulgare L.) under salt stress. Microbiol Res 181:22–32. https://doi.org/10.1016/j.micres.2015.08.002
Bourles A, Guentas L, Chalkiadakis E, Majorel C, Juillot F, Cavaloc Y, Burtet-Sarramegna V, Medevielle V, Jourand P, Amir H (2019) New caledonian ultramafic conditions structure the features of Curtobacterium citreum strains that play a role in plant adaptation. Can J Microbiol 65:880–894. https://doi.org/10.1139/cjm-2019-0283
Alishahi F, Alikhani HA, Khoshkholgh-Sima NA, Etesami H (2020) Mining the roots of various species of the halophyte Suaeda for halotolerant nitrogen-fixing endophytic bacteria with the potential for promoting plant growth. Int Microbiol 23:415–427. https://doi.org/10.1007/s10123-019-00115-y
Chauhan H, Bagyaraj DJ, Selvakumar G, Sundaram SP (2015) Novel plant growth promoting rhizobacteria—prospects and potential. Appl Soil Ecol 95:38–53. https://doi.org/10.1016/j.apsoil.2015.05.011
Cabria GLB, Argayosa VB, Lazaro JEH et al (2014) Draft genome sequence of haloalkaliphilic Exiguobacterium sp. strain AB2 from Manleluag Ophiolitic Spring, Philippines. Genome Announc 2:2–3. https://doi.org/10.1128/genomeA.00840-14.Copyright
Al-Mailem D, Eliyas M, Khanafer M, Radwan S (2014) Culture-dependent and culture-independent analysis of hydrocarbonoclastic microorganisms indigenous to hypersaline environments in Kuwait. Microb Ecol 67:1–9. https://doi.org/10.1007/s00248-014-0386-5
Kasana RC, Pandey CB (2018) Exiguobacterium: an overview of a versatile genus with potential in industry and agriculture. Crit Rev Biotechnol 38:141–156. https://doi.org/10.1080/07388551.2017.1312273
Giammarinaro P, Leroy S, Chacornac JP, Delmas J, Talon R (2005) Development of a new oligonucleotide array to identify staphylococcal strains at species level. J Clin Microbiol 43:3673–3680. https://doi.org/10.1128/JCM.43.8.3673-3680.2005
Máthé I, Borsodi AK, Tóth EM, Felföldi T, Jurecska L, Krett G, Kelemen Z, Elekes E, Barkács K, Márialigeti K (2014) Vertical physico-chemical gradients with distinct microbial communities in the hypersaline and heliothermal Lake Ursu (Sovata, Romania). Extremophiles 18:501–514. https://doi.org/10.1007/s00792-014-0633-1
Roohi A, Ahmed I, Khalid N et al (2014) Isolation and phylogenetic identification of halotolerant/halophilic bacteria from the salt mines of Karak, Pakistan. Int J Agric Biol 16:564–570
Akram MS, Shahid M, Tariq M, Azeem M, Javed MT, Saleem S, Riaz S (2016) Deciphering Staphylococcus sciuri SAT-17 mediated anti-oxidative defense mechanisms and growth modulations in salt stressed maize (Zea mays l.). Front Microbiol 7:1–14. https://doi.org/10.3389/fmicb.2016.00867
Ali B (2019) Functional and genetic diversity of bacteria associated with the surfaces of agronomic plants. Plants 8:91. https://doi.org/10.3390/plants8040091
Vásquez-Ponce F, Higuera-Llantén S, Pavlov MS, Marshall SH, Olivares-Pacheco J (2018) Phylogenetic MLSA and phenotypic analysis identification of three probable novel Pseudomonas species isolated on King George Island, South Shetland, Antarctica. Braz J Microbiol 49:695–702. https://doi.org/10.1016/j.bjm.2018.02.005
Venkateswar Reddy M, Mawatari Y, Onodera R, Nakamura Y, Yajima Y, Chang YC (2017) Polyhydroxyalkanoates (PHA) production from synthetic waste using Pseudomonas pseudoflava: PHA synthase enzyme activity analysis from P. pseudoflava and P. palleronii. Bioresour Technol 234:99–105. https://doi.org/10.1016/j.biortech.2017.03.008
Jia N, Du J, Ding MZ et al (2015) Genome sequence of Bacillus endophyticus and analysis of its companion mechanism in the Ketogulonigenium vulgare-Bacillus strain consortium. PLoS One 10:1–17. https://doi.org/10.1371/journal.pone.0135104
Akdoğan M, Çelik E (2018) Purification and characterization of polyhydroxyalkanoate (PHA) from a Bacillus megaterium strain using various dehydration techniques. J Chem Technol Biotechnol 93:2292–2298. https://doi.org/10.1002/jctb.5572
Kumar M, Sundaram S, Gnansounou E, Larroche C, Thakur IS (2018) Carbon dioxide capture, storage and production of biofuel and biomaterials by bacteria: a review. Bioresour Technol 247:1059–1068. https://doi.org/10.1016/j.biortech.2017.09.050
Chen X, Yin J, Ye J, Zhang H, Che X, Ma Y, Li M, Wu LP, Chen GQ (2017) Engineering Halomonas bluephagenesis TD01 for non-sterile production of poly(3-hydroxybutyrate-co-4-hydroxybutyrate). Bioresour Technol 244:534–541. https://doi.org/10.1016/j.biortech.2017.07.149
Coutinho de Paula F, Kakazu S, Bilia Chimello de Paula C et al (2019) Burkholderia glumae MA13: a newly isolated bacterial strain suitable for polyhydroxyalkanoate production from crude glycerol. Biocatal Agric Biotechnol 20:101268. https://doi.org/10.1016/j.bcab.2019.101268
Kumar P, Singh M, Mehariya S, Patel SKS, Lee JK, Kalia VC (2014) Ecobiotechnological approach for exploiting the abilities of Bacillus to produce co-polymer of polyhydroxyalkanoate. Indian J Microbiol 54:151–157. https://doi.org/10.1007/s12088-014-0457-9
Urbina L, Wongsirichot P, Corcuera MÁ, Gabilondo N, Eceiza A, Winterburn J, Retegi A (2018) Application of cider by-products for medium chain length polyhydroxyalkanoate production by Pseudomonas putida KT2440. Eur Polym J 108:1–9. https://doi.org/10.1016/j.eurpolymj.2018.08.020
Anderson AJ, Haywood GW, Williams DR, Dawes EA (1990) The production of polyhydroxyalkanoates from unrelated carbon sources. In: Novel biodegradable microbial polymers. Springer, Netherlands, pp 119–129. https://doi.org/10.1007/978-94-009-2129-0_11
Vidhyalakshmi R, Valli Nachiyar C, Narendra Kumar G, Sunkar S, Badsha I (2018) Production, characterization and emulsifying property of exopolysaccharide produced by marine isolate of Pseudomonas fluorescens. Biocatal Agric Biotechnol 16:320–325. https://doi.org/10.1016/j.bcab.2018.08.023
Passos da Silva D, Matwichuk ML, Townsend DO, Reichhardt C, Lamba D, Wozniak DJ, Parsek MR (2019) The Pseudomonas aeruginosa lectin LecB binds to the exopolysaccharide Psl and stabilizes the biofilm matrix. Nat Commun 10:1–11. https://doi.org/10.1038/s41467-019-10201-4
Abinaya M, Vaseeharan B, Divya M, Vijayakumar S, Govindarajan M, Alharbi NS, Khaled JM, al-anbr MN, Benelli G (2018) Structural characterization of Bacillus licheniformis Dahb1 exopolysaccharide—antimicrobial potential and larvicidal activity on malaria and Zika virus mosquito vectors. Environ Sci Pollut Res 25:18604–18619. https://doi.org/10.1007/s11356-018-2002-6
Hu X, Pang X, Wang PG, Chen M (2019) Isolation and characterization of an antioxidant exopolysaccharide produced by Bacillus sp. S-1 from Sichuan Pickles. Carbohydr Polym 204:9–16. https://doi.org/10.1016/j.carbpol.2018.09.069
Wang G, Zhu L, Yu B, Chen K, Liu B, Liu J, Qin G, Liu C, Liu H, Chen K (2016) Exopolysaccharide from Trichoderma pseudokoningii induces macrophage activation. Carbohydr Polym 149:112–120. https://doi.org/10.1016/j.carbpol.2016.04.093
Tripathi P (2007) Nitric oxide and the immune response. Indian J Biochem Biophys 44:310–319
Westbrook AW, Ren X, Moo-Young M, Chou CP (2018) Engineering of cell membrane to enhance heterologous production of hyaluronic acid in Bacillus subtilis. Biotechnol Bioeng 115:216–231. https://doi.org/10.1002/bit.26459
Funding
This work was supported by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) with scholarships to MPPP, JCR, and MAF. Fellowship from National Council for Scientific and Technological Development CNPq to LFS (309086/2018-3). Fapesp Grant (Fundação de Apoio à Pesquisa do Estado de São Paulo) to GP (2009/52665-4; 2010/51458-9).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Maria Paula Parada-Pinilla, Maria Alejandra Ferreira, Juan Camilo Roncallo, and Alexia Nathália Brígido Assef. The draft of the manuscript was written by Maria Paula Parada-Pinilla and Gabriel Padilla, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
The authors declare that they were in agreement and remain in agreement in this publication.
Consent for publication
The authors declare authorization of all participants for publication.
Code availability
Not applicable.
Additional information
Responsible Editor: Melissa Fontes Landell
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(PDF 579 kb).
Rights and permissions
About this article
Cite this article
Parada-Pinilla, M.P., Ferreira, M.A., Roncallo, J.C. et al. Biopolymer production by halotolerant bacteria isolated from Caatinga biome. Braz J Microbiol 52, 547–559 (2021). https://doi.org/10.1007/s42770-021-00426-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-021-00426-1