Abstract
Emerging diseases often originate from host shifts of introduced pests or pathogens. Genetic resistance of the host to such diseases might be limited or absent due to the lack of coevolutionary history. We review six examples of major disease outbreaks on native tree species caused by different introduced pests and pathogens that led to large ecological and economical losses. In all six cases, high tree mortality was observed in natural populations with some surviving individuals exhibiting varying levels of genetic resistance. The abundance and distribution of resistant individuals and the heritability of resistance traits varies substantially among the cases. While chestnut blight wilt combined with ink disease has virtually eliminated mature Castanea dentata trees from North America, other severe emerging diseases, such as the ash dieback, have left many surviving trees and genetic variation in resistance to such diseases has been documented. We argue that the evolutionary potential of tree species to respond to new emerging diseases should not be underestimated. However, the risk of increased levels of inbreeding and loss of genetic diversity caused by low population sizes is a major concern. Maintenance of broad genetic diversity is an important issue in conservation and forestry management. We expect that future research targeting the genetic background of biotic resistance towards emerging diseases and the role of endophytic communities in protecting trees will facilitate the informed and science-based guidance required to manage and maintain forests with high resilience. International cooperation on limiting disease spread and the provision of early invasive pest or pathogen detection systems are essential.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Forest tree species are threatened by current changes in their environment caused by climate change, overexploitation and fragmentation of their habitats, altered disturbance regimes, and by—often unintended—introductions of invasive species [1]. The expected magnitude and speed of climate change challenges the biotic and abiotic adaptations of plants, including forest tree species [2, 3]. The risk of reduced fitness due to poor climatic adaption is likely to be accompanied by an increased risk of serious biotic stress from already established, spreading or newly introduced pests and pathogens [4–6]. Several examples of the dramatic effects on forest health from sudden outbreaks of emerging pests and pathogens are well documented, e.g., Dutch elm disease (DED) (on Ulmus [7]), sudden oak death (on Quercus [8]), chestnut blight (on Castanea [9]), ash dieback (ADB) (on Fraxinus excelsior [10]), white pine blister rust (on Pinus [11]), and on the pest side emerald ash borer (EAB) (reviewed by Villari et al. [12] on Fraxinus), European gypsy moth on a variety of North American tree species (reviewed by Davidson et al. [13]), pinewood nematodes (PWNs) on Asian and European pines [14], and Hemlock woolly adelgids (HWA) on Eastern and Carolina hemlock [15]. Each of these diseases has spread rapidly and created economic and ecological loss within a few decades.
Tree species coevolve with many antagonist species that typically do not cause major damages. However, under certain circumstances, such as when exotic pests and pathogens are introduced to new areas, the impact on a novel host species can be severe. When an insect pest or pathogen attacks a tree, the effect of the infestation can range from highly damaging, and culminating in the host’s death, to complete immunity of the host to any caused damage. Several factors are important for the outcome including the following: (1) severity of pathogen pressure, (2) probability of initial establishment or repellence, (3) success of subsequent pathogen development, and (4) tolerance to tissue invasion. These factors have been recently reviewed by Ennos [16••] and will not be covered in the present article. In this review, we will refer to host resistance as “the collective heritable characteristics by which a plant species, race, clone, or individual may reduce the probability of successful utilization of that plant as a host by an insect (or pathogen) species, race, biotype or individual,” as defined by Beck [17].
A Battle on Unequal Terms?
Trees accumulate biomass and develop a large photosynthetic apparatus as they grow old providing attractive habitats for insects, fungi, and microorganisms. Most tree species host a diverse endophytic community including mutualists, commensalists, and parasites [18]. Although the actual mechanism of the individual interactions often remains unknown, a large number of positive associations in relation to plant defenses has been documented [19]. The presence of endophytes can enhance resistance to pathogens [19, 21] and beneficial microbes in the roots can improve induced resistance through priming [22]. However, during coevolution, endophytes can switch multiple times between a mutualistic and a pathogenic/parasitic lifestyle on their host [23]. Pests and pathogens thus coevolve with their host species mutually exerting and adapting to the other selection pressures. The coevolution of defense mechanisms and counter defenses can lead to an “arms race” between trees and their antagonists generating ever new response mechanisms [24]. Alternatively, negative frequency-dependent selection and temporally or spatially varying selection can favor the coexistence and maintenance of genetic variation in defense mechanisms (“trench-warfare” [25]). Genetic resistance to pests and pathogens can be due to major resistance genes (R genes), or polygenic adaptation [26, 27] that determine constitutive and induced defenses. Resistance mechanisms in forest trees have been recently reviewed by Telford et al. [28••].
Tree species share life history traits that drive and limit their tempo and mode of evolution (reviewed by Petit and Hampe [29]). The long generation time of trees provides a challenge for coadaptation since pests and pathogens usually have much shorter generation times and can evolve favorable traits during the lifespan of the host. However, trees also host mutualistic endophytes, mainly bacteria and fungi, with similarly short generation times as their pests and pathogens that can confer resistance [21, 30]. Furthermore, tree species typically maintain large effective population sizes with high-standing genetic variation [29] that are expected to allow for fast shifts in adaptive allele frequencies [31] if exposed to strong selection pressure. Newly introduced diseases can cause high mortality and thereby exert strong selection pressures favoring individual trees with low susceptibility [32••]. Individual tree death provides forest gaps where a dense regeneration can take place followed by selection among a large number of offspring when mortality is high. In this sense, evolutionary change is expected to occur much faster in response to severe pests and pathogens compared to altered climatic conditions where maladapted mature trees can still persist, although not thrive, for a long time [33].
Typically, introduced species are only able to become a threat on host species closely related to their native host [34], while the ability of the new host to cope with novel pests and pathogens depends on its own evolutionary history. In the worst case scenario, resistance to the new antagonist may be completely absent, leaving the host species without any potential for adaptation through natural selection on standing genetic variation. However, exaptations (sensu Gould and Vrba [35]) might confer resistance to invasive pests and pathogens although they coevolved in response to other selection pressures.
The objective of this paper is to review and discuss the adaptive potential of tree species to cope with novel insect pests and infectious diseases. Based on six case stories, which include some of the most severe epidemics on trees within the last century, we explore whether genetic variation in resistance was reported and the relative abundance and distribution of resistant individuals. Supported by the findings from the literature, we discuss the basis of the adaptive potential of trees to deal with new diseases. Finally, we discuss how conservation and landscape management of genetic diversity can support resilient forests in the next century.
Resistant Trees in Natural Populations: Lessons Learned from Twentieth Century Major Outbreaks
Dieback of European Ash
Natural populations of European ash (Fraxinus excelsior) have over the last two decades increasingly suffered damage due to ash dieback (ADB) caused by the invasive pathogenic fungus Hymenoscyphus fraxineus (Fig. 1). The introduction history is not fully clarified, but the first reports of disease symptoms came from Poland in the mid-1990s. It is likely that the pathogen was introduced through movement of Fraxinus mandshurica plants from Asia to Eastern Europe that led to a host shift to F. excelsior [36]. The disease has subsequently spread rapidly across Europe with the first reports of ash dieback symptoms in Scandinavia around 2001 and most recently in UK in 2012 [32••]. The disease causes substantial mortality, especially in young populations [37] and the abundance of healthy individuals was found to be low in most infected areas. In Denmark, less than 5 % of more than 6000 trees in two test plantings with offspring from trees of local origin remained healthy 10 years after planting [32••]. Husson et al. [38] found only 8 % healthy trees in a large survey of 2400 trees across 60 forest plots in France. However, the presence of genetic variation in resistance was confirmed from several countries [39–46] with moderate to high levels of heritability (h 2) and genetic coefficient of variation (CVg) for susceptibility, 0.1–0.6 for CVg, and 0.3–0.6 for h 2, respectively [39–44]. Based on breeding value estimates, the frequency of genotypes with high resistance is expected to be relatively low (1–5 % or less [32••]) but it is interesting that genetic variation in resistance was reported in all the studied populations indicating that European ash has the potential to undergo rapid evolution towards higher levels of resistance through natural or artificial selection. Although 1–5 % is a low frequency, it implies that trees with high levels of resistance are expected to be present in almost any native ash forest and selection in favor of increased resistance is likely to be ongoing. Pliūra et al. [42] found in a provenance study that ash offspring from trees from Baltic countries were significantly less affected by the disease compared to offspring from trees from the Western part of the natural distribution area when grown under the same conditions in Lithuania. Given the disease history, with the Baltic countries being the epicenter for the first symptoms and high mortality, these observed differences between Western and Eastern populations could reflect an already realized response to natural selection.
Damages caused by Hymenoscyphus fraxineus on Fraxinus excelsior. a Variation in degree of crown damage among infested trees in a planted stand in Denmark. b Lesion on a European ash leaflet after controlled inoculation. c Fruiting bodies of H. fraxineus on leaf rachises and petioles. d Lesion on a young stem after controlled inoculation with an infested wood plug. Photos: Lars N. Hansen, Lene R. Nielsen, and Lea Vig McKinney
Ink Disease and Chestnut Blight
American chestnut (Castanea dentata) was an important tree species in North East American forests until it was heavily decimated across its native range during the last century by the combined effect of Phytophthora cinnamomi causing ink disease and Cryphonectria parasitica causing chestnut blight.
Phytophthora cinnamomi was probably introduced to the native range of American chestnut two centuries ago and Cr. parasitica a century later [47]. Therefore, the two pathogens have been present in NE American ecosystems for a long time. Some chestnut trees have been able to survive by resprouting [9] and a few old surviving trees were observed to exhibit some degree of resistance. These candidates might be good candidates for breeding [48, 49•]. Alexander et al. [50] reported that old healthy trees are rare and difficult to find, and Hebard [51] expects that very few mature trees (DBH >33 cm) have survived in the core of the distribution area. The variation in health among trees has been found to be influenced by the virulence of Cr. parasitica-specific strains, because hypovirulent pathogen strains infected with the RNA virus do not kill the trees. The interaction is complicated since the susceptibility of the trees seems to depend on the interaction between the tree genotype, its growing conditions, and the virulence of the Cr. parasitica strain [52]. The chestnut blight is one of the most investigated emerging infectious diseases on trees and substantial genomic resources have been developed recently. However, information on the frequency of resistant trees in the natural forests or on levels and presence of additive genetic variation based on progeny trials is very limited. Quantified estimates of intraspecific variation in disease resistance is probably lacking due to a focus on hybridization with Asian species to increase resistance since American chestnut was early recognized as highly susceptible [53]. The apparently very low frequency of mature trees that have survived the disease in the native habitat and a lack of healthy recruitment from such rare survivors suggest that the potential is limited at least on a short timescale. Mature trees do still exist and some of these may have high natural resistance, but estimates of heritability are, to our knowledge, unknown. Recent activities have included development of genetically modified clones [54] as a potential option for increasing resistance, but here the approach is based on genes not already present in the species.
Hybridization with Asian chestnut species has also been deployed to obtain resistance towards Ph. cinnamomi in Europe. However, natural resistance has been observed recently among pure European chestnut (Castanea sativa) trees. In a European study testing 50 clones of various origins, one natural European chestnut genotype was as resistant as the hybrid used as a resistant control [55]. A very large screening across thousands of hectares in Spain led to the identification of 209 Ca. sativa trees of which more than 100 were propagated and subjected to thorough screening. Two of these clones were classified as resistant and three clones at least partly resistant [56]. These two studies were conducted on clones so additive genetic variation could not be estimated for resistance. However, the existence of rare resistant genotypes embedded in the large gene pool of chestnut trees that were susceptible to the introduced pathogen supports the expectation that evolution towards increased resistance over time can take place. The low abundance of resistant trees in a large area is of concern because genetic bottlenecks and inbreeding could decrease genetic diversity unless very effective gene flow counteracts the effects of small population sizes.
Phytophthora cinnamomi is a pathogen that infests and causes serious damage in a large number of tree species across families and genera [57]. Frampton et al. [58] observed substantial genetic variation in susceptibility of the two closely related Abies equi-trojani and Abies bornmuelleriana species in Turkey, where Ph. cinnamomi is also considered to have been introduced. Based on controlled infections on offspring from single tree collections covering the native range of the two Abies species in Turkey, moderate to high narrow sense heritabilities (i.e., reflecting additive genetic effects) for resistance of 0.5–0.6 were estimated. The authors also observed variation in the level of susceptibility among populations revealing an interesting east–west gradient. The background behind this gradient is unknown but made the authors speculate that genetic variation in resistance to the introduced Ph. cinnamomi pathogen could be due to exaptation due to adaptation to other Phytophthora species present in the region [58].
Ulmus and DED
The effect of Dutch elm disease (DED) represents another example of a major calamity caused by an emerging infectious disease. Two major outbreaks caused by the pathogenic fungi Ophiostoma ulmi and Ophiostoma novo-ulmi have led to the death of millions of European elm trees during the last century [59]. The density of large elm trees has severely decreased across Europe but occasional mature trees are still found in the landscape. Young seedlings of Ulmus glabra are commonly observed in at least parts of the natural distribution area [60]. The second wave of the disease caused by O. novo-ulmi created very high mortality [61] and breeding for resistance towards the new disease soon focused on introducing resistance through hybridization with Asian Ulmus species [62, 63]. However, gene conservation strategies, based on an in situ conservation approach, have been developed to utilize the natural populations’ ability to respond to the selection pressure imposed by the pathogen [64]. Also, breeding programs have been initiated to develop resistant planting material of the pure Ulmus minor in Spain. Unfortunately, the breeding efforts were hampered by the small fraction (0.5 %) of the trees showing resistance [65], but, based on selection and testing at multiple sites, seven U. minor clones tolerant to O. novo-ulmi were identified and released for use in reforestation efforts [66]. Venturas et al. [67] reported moderate to high narrow-sense heritability (h 2 = 0.54) in U. minor after controlled inoculations. Although the frequency of trees with high levels of resistance was low, the presence of genetic variation and moderate to high heritability suggest that significant evolutionary potential is still present in the natural populations of elms in Europe. Similar results have been obtained in North America, where selection and testing have identified genotypes of American elm (Ulmus americana) with very low susceptibility [68] and successful breeding programs have been implemented [49•].
The Emerald Ash Borer
The emerald ash borer (EAB, Agrilus planipennis) was most likely introduced to North America in the 1990s [69] but was not recognized as a new forest pest until 2002 [70]. It originates from eastern Asia [71] but has already colonized ample areas in North America and killed millions of ash trees while it continues to spread [72]. White (Fraxinus americana), green (Fraxinus pennsylvanica), and black (Fraxinus nigra) ash are widespread, important forest components in North America and are highly susceptible to EAB [72, 73]. The larvae feed on the phloem and trees usually die 3–4 years after infestation, young trees even earlier [72]. The percentage of mortality in natural populations can exceed 99 % in highly infested stands in Michigan and regeneration is extremely limited [74]. So far, all North American ash species in contact with EAB seem to be susceptible [73, 75, 76] though blue ash (Fraxinus quadrangulata) to a lower degree [77]. Asian ash species, especially Fraxinus mandshurica show distinct, induced, and constitutive phloem chemistry and appear more resistant to EAB, mainly because female EAB avoid healthy trees for oviposition (reviewed by Villari et al. [12]). However, in North America, so far, only a few genotypes per species have been tested for resistance to EAB in studies targeting interspecific variation [73, 75, 76] and an extensive screening of more genotypes for intraspecific variation is essential in the future [12].
Surviving ash trees in heavily infested natural stands exist—although they are rare—(<0.1 %) [74], and these are likely promising candidates that should be tested for resistance in controlled conditions [78] and eventually used as resource for resistance in breeding programs [12]. A first bioassay study reported different mechanisms of resistance in these “lingering” ashes for F. pennsylvanica, and a breeding program to increase resistance in this species based on these trees has been implemented [79].
Recently, EAB has also been reported to cause damage in ash trees (Fraxinus spp.) in urban areas in Moscow, Russia [80]. In forests south of Moscow, widespread trees of European ash (F. excelsior) have also been infested and suffer dieback although they seem to be less susceptible than North American species [81]. Research evaluating intraspecific resistance of the three European ash species to EAB is pressing since the beetle is most likely to spread in Europe [81].
Hemlock Woolly Adelgids
The hemlock woolly adelgid (HWA, Adelges tsugae) was introduced to North America in the 1950s from southern Japan [82]. Over the last few decades, the species has invaded vast areas in North America where it caused extensive mortality of eastern (Tsuga canadensis [83]) and Carolina hemlock (Tsuga caroliniana [84]). The damage in northeastern USA has been severe with adelgid-induced mortality exceeding 95 % and 50–75 % defoliation in surviving trees [85, 86]. Attacked trees stop their growth, drop attacked needles, and usually die 4–10 years after infestation [87]. Natural regeneration after HWA infestation is rare because affected trees do not produce seeds and are unable to re-sprout [85]. Forest management employing biological control agents such as Sasajiscymnus tsugae or Laricobius nigrinus have reduced HWA density only locally [88] and preventive salvage logging has additionally aggravated the impact [89]. In many regions, previously hemlock-dominated forests underwent a severe change in species composition after HWA attack [89, 90]. In contrast Asian and even western North American hemlock species seem to be resistant to HWA [91]. Not surprisingly, mitochondrial DNA studies revealed a long coevolutionary history between HWA and hemlock species in western North America and Asia [82]. Recently, few individual eastern hemlock trees with resistance to HWA were also found [92, 93]. Nutritional foliar chemistry [94], as well as terpenoid abundance [95], might be involved in this lower susceptibility. The future impact of HWA is expected to be exacerbated by climate change since warmer winters are expected to enable an unimpeded spread of this pest also in the northern distribution range of eastern American hemlock species [96]. Knowledge about the abundance and distribution of resistant trees and the narrow sense heritability of the trait is to our knowledge so far lacking.
Pinewood Nematode
Bursaphelenchus xylophilus, the pinewood nematode (PWN) causes the pine wilt disease and is a serious threat especially but not limited to pine populations. The species is native to North America but was introduced to Asia in the early twentieth century where it caused a severe dieback in pine populations [97]. In the 1980s, the species spread from Japan to China and Korea (reviewed by Zhao et al. [98]) and at the end of the 1990s, it was brought to Portugal [99]. The Portuguese government implemented the National Eradication Program for the Pinewood Nematode (PROLUNP), which aimed to log all symptomatic trees in order to avoid further spread of the disease [100]. The program was carried out in a small affected area and a demarcation area surrounding it. When newly infested trees were detected in the demarcation area, the limits were redefined, and a clear-cut corridor (3-km wide), free of all tree species that could potentially host PWN, was prepared. Despite these efforts, PWN spread quickly in Pinus pinaster forests causing sudden wilt and tree death [101]. The disease is mainly spread by the movement of forest products [101] but in nature, it is also dispersed by its vector species, a beetle of the genus Monochamus [102]. Apart from logging of infested trees, forest management against the pinewood nematode in Asia includes large-scale insecticide spraying from helicopters or planes to prevent the spread of the vector species [14].
Infested trees usually die 40 days after infestation [103]. Some resistant trees have been observed and studied in the usually susceptible Pinus thunbergii and Pinus densiflora [104, 105]. In the 1970s, breeding programs for resistance were initiated based on resistant cultivars, and seedlings from the program have been used for reforestation since the 1990s [106]. European pine species show interspecific differences in susceptibility to PWN with Pi. pinaster being the most susceptible and Pinus pinea as the most resistant species [107]. However, Zas et al. [108] reported intraspecific variation in resistance to PWN in Pi. pinaster at the provenance level. These findings suggest that genetic variation might be present and more genetic studies targeting resistance to PWN are needed. The expression of the disease is related to temperature [109], because the nematodes can be present in trees without causing symptoms when summer temperatures are low. With global warming, the disease might spread further north in Europe where Pinus sylvestris dominates in the extensive Boreal forests, and is considered a very susceptible host [110].
The Origin of Genetic Variation in Resistance Towards New Emerging Pests and Pathogens
Where Does the Variation in Resistance Come From?
Phenological mismatch between the pathogen and its new host may result in disease escape of host genotypes in the extreme ends of the natural variation in growth rhythm. In the case of ash dieback, McKinney et al. [39] observed a strong genetic correlation (rg > 0.7) between resistance and early autumn leaf coloration. Since the fungus infects the host through the leaves during summer, early leaf senescence might increase the probability of disease escape [39]. A correlation between phenology and susceptibility was also reported for Ulmus species affected by DED. Here, early flushing may indicate disease avoidance due to a phenological/physiological mismatch with the occurrence of the pathogen vector (species of the genus Scolytus which feed and breed under the bark) [111, 112]. Furthermore, smaller and narrower vessels seemed to limit pathogen growth [67] and standing variation in these anatomical traits may, therefore, explain part of the variation in susceptibility. In sudden oak death canker disease, the pathogen Phytophthora ramorum sporulates early in the year and requires active host cambial tissue to successfully infect Quercus sp. Therefore, late-flushing host trees can escape the disease [113]. Genetic variation in phenology is maintained in natural populations when selection pressures shift between years, likely due to annual variation in occurrence of spring or autumn frosts. This variation can serve as buffer against damage from newly introduced pathogens or pests.
Most plant species have experienced and coevolved with a large number of interacting organisms during their long evolutionary history and range shifts. Therefore, genetic variation in host resistance might have arisen during time slots of exposure to this or similar pests and pathogens. Random genetic drift is a weak force if effective population sizes are large [114], and genetic variation from previous exposure to similar pathogens can, therefore, have been maintained at a low frequency in natural populations even in the absence of selection. The ash die back pathogen H. fraxineus is closely related to a native European fungus Hymenoscyphus albidus, which is considered a harmless decomposer of leaves from European ash. It has been speculated that this relationship may have previously involved a degree of pathogenicity and adaptive polymorphisms that evolved under previous selection pressures and might, therefore, remain in the gene pool of the host species [41]. Along the same line, a large population decline in European elms, as indicated by pollen diagrams from approximately 5000–6000 BP, has been proposed to have resulted from an epidemic spread by Scolytus species. This may have been similar to the outbreak of DED in the past century [115]. It can, therefore, be speculated that such an outbreak may have generated exaptation in the host.
Another important aspect of the evolutionary potential of tree populations to cope with emerging diseases is their associated endophytic community, mainly fungi and bacteria [4]. Recently, endophyte communities have been proposed as indicators of tree health [116]. Gennaro et al. [117] found the endophytic communities on declining oaks infected by oak puzzle disease to be less diverse than those on healthy trees. Tubakia dryina was found more often on diseased trees while Monochaetia monochaeta was more abundant on healthy trees. In contrast, Martin et al. [118] found that U. minor genotypes with resistance against O. novo-ulmi (DED) had lower frequency and diversity of fungal endophytes in the xylem than susceptible U. minor genotypes. However, in laboratory conditions, Díaz et al. [119] demonstrated that an isolate of Trichoderma atroviride extracted from elm trees inhibits growth of O. novo-ulmi and was, therefore, proposed to confer resistance to DED. In Populus, several endophytic species seemed to contribute to quantitative resistance to Melampsora rust [20]. The most commonly reported role of endophytes is a strong induced resistance response in hosts due to previous contact with an endophyte. For example, Pinus monticola seedlings were more resistant to white pine blister rust if they had been previously exposed to endophytes [30]. Arnold et al. [21] also showed that inoculation of endophyte-free leaves of Theobroma cacao with endophytes from naturally infected, asymptomatic trees could reduce leaf mortality of seedlings exposed to Phytophthora sp. The increased defense was primarily localized in the endophyte-infected tissues. The use of endophytes as biological control agents to manage forest diseases has been recently discussed [120•]. However, the community composition and role of endophytes in tree disease resistance, especially under natural conditions, remains poorly understood and harbors a promising field of research opportunities.
Implication for Forest Management and Conservation
Most emerging infectious diseases and destructive insects are caused by accidental introductions [115]. Therefore, obvious precautions include the limitation, or diligent control, of long-distance transfer of plant material and products that can serve as vectors for unintended introduction of insects, fungi, or other microorganisms (see, e.g., Montesclaros declaration [121]). Moreover, the development of international cooperation for disease management is essential [122]. The probability of a successful host shift from an introduced species is low, but most cases of successful establishment on a new host leads to dramatic and largely negative consequences.
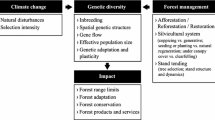
Do Not Underestimate the Evolutionary Potential of Tree Species but Reduce the Risk of Genetic Bottlenecks
This review of six major emerging diseases from the last century highlights the evolutionary potential of natural tree populations to respond to completely new pathogenic species. A common picture from these study cases is that the number of individual trees has been dramatically and rapidly reduced following the emergence of the new disease, but the species were not eradicated. Survival can be due to either disease escape or to different resistance mechanisms that are at least partly under genetic control and harbor moderate-to-high narrow sense heritability. This low abundance of unaffected trees, which are often scattered over large areas, can lead to severe genetic bottlenecks. The limited access to pollen from conspecific individuals may result in increased self-pollinations, increased relatedness among offspring in a given area, and decreased intraspecific genetic diversity. However, Nielsen and Kjær [123] studied surviving, scattered and solitary wind pollinated elm trees in the Danish landscape after the DED outbreak and found no genetic effects in relation to the lowered tree density. The offspring from these surviving trees were outcrossed, genetically diverse, and progeny from the same mother tree had been sired mostly by several different pollen donors. Long pollination distances have also been reported by Bacles and Ennos [124] in the wind pollinated ash (F. excelsior) in a fragmented landscape prior to ash dieback. Wind-pollinated conifers are also expected to maintain very large effective population sizes through pollen flow over long distances.
Nevertheless, the potential negative effects of forest fragmentation should be taken seriously [125]. Since trees with sufficient genetic resistance may be <1 % in natural populations, the risk of decreased fitness due to inbreeding depression is a serious concern, especially in insect-pollinated trees and low abundance species growing in mixed forests. This can develop into a negative feedback loop, if loss of vigor leads to replacement by other species, which again reduces the effective population size and limits seed dispersed for next generation recruits. Silviculture in support of these endangered species may, therefore, be important. Unmanaged naturalized forests are expected to be less affected than forest plantations and resilience in these forests can be further supported through the maintenance of large population sizes, and should involve long-term land use planning to ensure continuous forest patches where gene flow can take place at the landscape level.
Resistant, but Also Genetically Diverse Seed Sources for Reforestation
Maintenance of substantial genetic variation in seed destined for planting programs of trees in long rotation will support the adaptive potential of planted forests and ensure a low a priori relatedness among planted trees [126]. On the contrary, high-genetic homogeneity has been suggested to facilitate a fast disease spread, e.g., Gil et al. [127] speculated that the spread of DED on English elm in Britain was promoted by scattered plantings and vegetative reproduction of a single clone during centuries. Breeding programs must identify and test a substantial number of surviving and healthy trees based on large-scale screening to be an effective tool for the development of genetically diverse and disease-resistant seed sources for forest restoration [49]. For example, the Danish restoration program for ash forests includes the testing of more than 200 trees selected among thousands of trees across the Danish landscape and similar programs are being initiated in other countries [32]. The public is very concerned about forest health, and involving citizen science is an interesting option for the identification and continuous monitoring of surviving trees [128, 129]. This approach can multiply tenfold the identified number of healthy trees, as it is time consuming and expensive to find rare healthy trees scattered across large forest areas without the help of local people. Besides finding trees to be included in breeding programs for restoration, these observations from citizen science can improve data quality on disease spread. Also, local or regional efforts to protect these surviving trees can be implemented more broadly, efficiently, and effectively. Volunteers have already been involved in monitoring tree health in forests suffering from ash dieback in the UK (http://www.observatree.org.uk/portal/tree-health-citizen-science-projects) and sudden oak death in the USA (http://oakmapper.org/).
Felling of Healthy Trees in the Neighborhood of Diseased Areas can be Counterproductive on a Large Scale
A classical forest management tool to avoid the spread of new pests and diseases is preemptive and salvage logging in the neighborhood of an infested forest patch. This strategy is highly relevant at the very beginning when a newly introduced pest or pathogen is detected for the first time. It should be employed in initial and locally restricted cases of first-disease incidence. However, as soon as several disease centers are emerging, it can become counterproductive due to the removal of high numbers of healthy and some potentially resistant trees. Since healthy mature forest trees represent commercial value to the forest land owner, the outbreak of a new infectious disease could lead to the extensive logging of many healthy trees to minimize the risk of lost revenue due to infections [32••]. In this way, a new disease can trigger both natural mortality that will reduce the density of susceptible trees and increase harvesting that will further decrease the density of all trees including the rare resistant ones. In some cases, this strategy has been successful, e.g., Asian long-horned beetles were eradicated in Illinois and Jersey City after an initial introduction [130]. In other cases, preemptive logging has not proven successful, e.g., the spread of PWN in Portugal 1999–2009 [131] and of the EAB in Canada [132] could not be avoided. However, more research is needed on this topic since few studies have addressed the effectiveness and impacts of preemptive logging so far. The usefulness of this management tool is highly case-specific and depends e.g., on the mode of disease spread. Foster et al. [133] also pointed out that preemptive/salvage logging often imposes a bigger ecosystem impact than the disturbance itself.
Cooperation and Early Warning Systems Based on Observations in Arboreta
Arboreta and plantings of exotic tree species can inform about potential risks of pests and pathogens before they are accidentally introduced to other jurisdictions. For example, an arboretum was used to study interspecific variation in the susceptibility to HWA among Tsuga species from different continents [134]. There are ongoing initiatives such as COST action FP1401 “A global network of nurseries as early warning system against alien tree pests (Global warning)” [135] or the project “REINFFORCE - REsource INFrastructure for monitoring and adapting European Atlantic FORests under Changing climate” [136] that use arboreta to detect possible future biotic threats. Furthermore, experimental plantations of Norway spruce (Picea abies) in North America suffered severe damage from the native white pine weevil (Pissodes strobi), an insect pest that naturally co-occurs and feeds on several North American conifer species. Within these plantations, Norway spruce revealed a degree of susceptibility similar to highly damaged local Sitka spruce populations. In an Estonian arboretum, Drenkhan et al. [137] screened exotic Fraxinus species for resistance to the ash dieback pathogen and observed signs of infection on Fraxinus species native to areas in North America where the pathogen is not present.
Implication for Gene Conservation Programs
Conservation of genetic resources of key forest tree species is recognized as an important part of sustainable forest management [138] and various guidelines have been developed that typically target minimum effective population sizes of 50–5000 [139]. In general, these numbers are derived from “the golden rule of 50–5000,” which remains controversial and hotly debated in conservation literature [140]. However, if an emerging disease creates high mortality leaving only 1 % living trees scattered across populations, a requirement for an effective population size of >50–5000 in the next generation will obviously require an initial gene conservation population where 5000 mature trees represent an absolute minimum. The effective population size is typically less than half the census number of mature trees in the landscape due to variation in fecundity; therefore, the actual number of mature trees that need to be conserved is larger than often anticipated [141] Also, since resistance may be related to interactions with the endophytic society in the trees, in situ conservation or in situ-like conservation approaches [142] may have a clear preference to ex situ programs, which are mainly concerned with conserving the genetic variation of the targeted tree species.
Conclusion
Emerging exotic pests and pathogens pose a major challenge for future global forests and should be addressed by international cooperation reducing the risk of new introductions. However, natural populations often exhibit some level of genetic resistance even to newly introduced species. The surviving trees may be scattered and limited in number, but the evolutionary potential of host species to cope with emerging pests and pathogens should not be underestimated. Natural resistance can be facilitated by maintaining genetic diversity in natural forests and by supporting connection between trees and forests to allow continuous gene flow. It will be a challenge to maintain viable natural population sizes of species that undergo dramatic mortality caused by an aggressive emerging disease. In managed forests, large-scale deployment of planting material with low genetic diversity can be extremely risky. Therefore, we suggest that breeding for resistance should not lead to genetically uniform populations with low resilience and small effective population sizes. Instead, breeding should involve large-scale screening efforts across natural populations to ensure that a high number of resistant trees are identified and included in the testing and breeding activities. Programs for the identification, protection, and propagation of surviving trees will be imperative and collaboration between scientists, forest managers, public authorities, and civil society will be essential. Future research on the genetics of resistance mechanisms, host-pathogen interactions, exaptation, and the role of endophytes will, hopefully, lead to a better understanding of the biotic adaptation progress that can guide effective forest management, disease control, resistance breeding, and restoration efforts.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Dale VH, Joyce LA, McNulty S, Neilson RP, Ayres MP, Flannigan MD, et al. Climate change and forest disturbances. Bioscience. 2001;51:723–34.
Sexton JP, Hangartner SB, Hoffmann AA. Genetic isolation by environment or distance: which pattern of gene flow is most common? Evolution. 2014;68:1–15.
Hoffmann AA, Sgrò CM. Climate change and evolutionary adaptation. Nature. 2011;470:479–85.
Pautasso M, Schlegel M, Holdenrieder O. Forest health in a changing world. Microb Ecol. 2015;69:826–42.
Boyd IL, Freer-Smith PH, Gilligan CA, Godfray HCJ. The consequence of tree pests and diseases for ecosystem services. Science. 2013;342:1235773.
Santini A, Ghelardini L, De Pace C, Desprez-Loustau ML, Capretti P, Chandelier A, et al. Biogeographical patterns and determinants of invasion by forest pathogens in Europe. New Phytol. 2013;197:238–50.
Brasier C. Intercontinental spread and continuing evolution of the Dutch elm disease pathogens. In: Dunn C, editor. Elms. US: Springer; 2000. p. 61–72.
Rizzo DM, Garbelotto M, Hansen EM. Phytophthora ramorum: integrative research and management of an emerging pathogen in California and Oregon forests. Annu Rev Phytopathol. 2005;43:309–35.
Elliott K, Swank W. Long-term changes in forest composition and diversity following early logging (1919–1923) and the decline of American chestnut (Castanea dentata). Plant Ecol Springer Netherlands. 2008;197:155–72.
Pautasso M, Aas G, Queloz V, Holdenrieder O. European ash (Fraxinus excelsior) dieback—a conservation biology challenge. Biol Conserv. 2013;158:37–49.
Maloy OC. White pine blister rust control in North America: a case history. Annu Rev Phytopathol. 1997;35:87–109.
Villari C, Herms DA, Whitehill JGA, Cipollini D, Bonello P. Progress and gaps in understanding mechanisms of ash tree resistance to emerald ash borer, a model for wood-boring insects that kill angiosperms. New Phytol. 2015; doi: 10.1111/nph.13604
Davidson CB, Gottschalk KW, Johnson JE. Tree mortality following defoliation by the European Gypsy Moth (Lymantria dispar L) in the United States: Annu. Rev For Sci. 2006;45:74–84.
Mota M, Vieira P. Pine wilt disease a worldwide threat to forest ecosystems. Springer Netherlands. 2008. 405 pp
Kizlinski ML, Orwig DA, Cobb RC, Foster DR. Direct and indirect ecosystem consequences of an invasive pest on forests dominated by eastern hemlock. J Biogeogr. 2002;29:1489–503.
Ennos R. Resilience of forests to pathogens: an evolutionary ecology perspective. Forestry. 2014;88:41–52. This paper reviews the factors that determine the levels of pathogen damage experienced in forest trees and the conditions which enable or preclude a stable co-existence of host and pathogen.
Beck S. Resistance of plants to insects. Annu Rev Entomol. 1965;10:207–32.
Sieber TN. Endophytic fungi in forest trees: are they mutualists? Fungal Biol Rev. 2007;21:75–89.
Hardoim PR, van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A, et al. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev. 2015;79:293–320.
Raghavendra AKH, Newcombe G. The contribution of foliar endophytes to quantitative resistance to Melampsora rust. New Phytol. 2013;197:909–18.
Arnold AE, Mejía LC, Kyllo D, Rojas EI, Maynard Z, Robbins N, et al. Fungal endophytes limit pathogen damage in a tropical tree. Proc Natl Acad Sci. 2003;100:15649–54.
Conrath U, Thulke O, Katz V, Schwindling S, Kohler A. Priming as a mechanism in induced systemic resistance of plants. Eur J Plant Pathol. 2001;107:113–9.
Delaye L, García-Guzmán G, Heil M. Endophytes versus biotrophic and necrotrophic pathogens-are fungal lifestyles evolutionarily stable traits? Fungal Divers. 2013;60:125–35.
Dawkins R, Krebs JR. Arms races between and within species. Proc. R. Soc. Lond. B. Biol. Sci. 1979;205:489–511.
De Meaux J, Mitchell-Olds T. Evolution of plant resistance at the molecular level: ecological context of species interactions. Heredity. 2003;91:345–52.
Gebhardt C, Valkonen JP. Organization of genes controlling disease resistance in the potato genome. Annu Rev Phytopathol. 2001;39:79–102.
Carson SD, Carson MJ. Breeding for resistance in forest trees—a quantitative genetic approach. Annu Rev Phytopathol. 1989;27:373–95.
Telford A, Cavers S, Ennos RA, Cottrell JE. Can we protect forests by harnessing variation in resistance to pests and pathogens? Forestry. 2014;88:3-12. This paper reviews and explains the resistance mechanisms in trees and the effect of genetical and environmental factors on resistance trait variation.
Petit RJ, Hampe A. Some evolutionary consequences of being a tree. Annu Rev Ecol Evol Syst. 2006;37:187–214.
Ganley RJ, Sniezko RA, Newcombe G. Endophyte-mediated resistance against white pine blister rust in Pinus monticola. For Ecol Manage. 2008;255:2751–60.
Barrett RDH, Schluter D. Adaptation from standing genetic variation. Trends Ecol Evol. 2008;23:38–44.
McKinney L V, Nielsen LR, Collinge DB, Thomsen IM, Hansen JK, Kjær ED. The ash dieback crisis: genetic variation in resistance can prove a long-term solution. Plant Pathol. 2014;63:485–499. This paper reviews the subsequent spread of ash dieback in Europe and points to the importance of naturally occurring resistance to non-native pathogens as basis for natural or assisted selection.
Kuparinen A, Savolainen O, Schurr FM. Increased mortality can promote evolutionary adaptation of forest trees to climate change. For Ecol Manage. 2010;259:1003–8.
Gilbert GS, Webb CO. Phylogenetic signal in plant pathogen-host range. Proc Natl Acad Sci. 2007;104:4979–83.
Gould S, Vrba E. Exaptation—a missing term in the science of form. Paleobiology. 1982;8:4–15.
Drenkhan R, Sander H, Hanso M. Introduction of Mandshurian ash (Fraxinus mandshurica Rupr.) to Estonia: is it related to the current epidemic on European ash (F. excelsior L.)? Eur J For Res. 2014;133:769–81.
Lobo A, Hansen JK, McKinney LV, Nielsen LR, Kjær ED. Genetic variation in dieback resistance: growth and survival of Fraxinus excelsior under the influence of Hymenoscyphus pseudoalbidus. Scand J For Res. 2014;29:519–26.
Husson C, Caël O, Grandjean JP, Nageleisen LM, Marçais B. Occurrence of Hymenoscyphus pseudoalbidus on infected ash logs. Plant Pathol. 2012;61:889–95.
McKinney LV, Nielsen LR, Hansen JK, Kjær ED. Presence of natural genetic resistance in Fraxinus excelsior (Oleraceae) to Chalara fraxinea (Ascomycota): an emerging infectious disease. Heredity. 2011;106:788–97.
McKinney LV, Thomsen IM, Kjær ED, Bengtsson SBK, Nielsen LR. Rapid invasion by an aggressive pathogenic fungus (Hymenoscyphus pseudoalbidus) replaces a native decomposer (Hymenoscyphus albidus): a case of local cryptic extinction? Fungal Ecol. 2012;5:663–9.
Kjær ED, McKinney LV, Nielsen LR, Hansen LN, Hansen JK. Adaptive potential of ash (Fraxinus excelsior) populations against the novel emerging pathogen Hymenoscyphus pseudoalbidus. Evol Appl. 2012;5:219–28.
Pliūra A, Lygis V, Suchockas V, Bartkevicius E. Performance of twenty four European Fraxinus excelsior populations in three Lithuanian progeny trials with a special emphasis on resistance to Chalara fraxinea. Balt For. 2011;17:17–34.
Pliūra A, Lygis V, Marciulyniene D, Suchockas V, et al. Genetic variation of Fraxinus excelsior half-sib families in response to ash dieback disease following simulated spring frost and summer drought treatments. iForest - Biogeosciences For. 2015; doi: 10.3832/ifor1514-008
Lobo A, McKinney LV, Hansen JK, Kjær ED, Nielsen LR. Genetic variation in dieback resistance in Fraxinus excelsior confirmed by progeny inoculation assay. For. Pathol. 2015; doi: 10.1111/efp.12179
Stener LG. Clonal differences in susceptibility to the dieback of Fraxinus excelsior in southern Sweden. Scand J For Res. 2013;28:205–16.
Kirisits T, Freinschlag C. Ash dieback caused by Hymenoscyphus pseudoalbidus in a seed plantation of Fraxinus excelsior in Austria. J Agric Ext Rural Dev. 2012;4:184–91.
Clark SL, Schlarbaum SE, Pinchot CC, Anagnostakis SL, Saunders MR, Thomas-Van Gundy M, et al. Reintroduction of American chestnut in the National Forest System. J For. 2014;112:502–12.
Griffin GJ. Blight control and restoration of the American Chestnut. J For. 2000;98:22–7.
Sniezko RA. Resistance breeding against nonnative pathogens in forest trees—current successes in North America. Can J Plant Pathol. 2006;28:S270–9. This paper highlights the importance of naturally occurring resistant trees in forests threatened by non-native pests and pathogens and provides examples of successful breeding programs in North America.
Alexander MT, Worthen LM, Craddock JH. Conservation of Castanea dentata germplasm of the southeastern United States. 693rd ed. Leuven, Belgium: International Society for Horticultural Science (ISHS); 2005. p. 485–90.
Hebard FV, Fitzsimmons SF, Gurney KM, Saielli TM. The breeding program of the American Chestnut Foundation. 1019th ed. Leuven, Belgium: International Society for Horticultural Science (ISHS); 2014. p. 135–9.
Milgroom MG, Cortesi P. Biological control of Chestnut blight with hypervirulence: a critical analysis. Annu Rev Phytopathol. 2004;42:311–38.
Anagnostakis SL. Chestnut breeding in the United States for disease and insect resistance. Plant Dis. 2012;96:1392–403.
Merkle SA, Andrade GM, Nairn CJ, Powell WA, Maynard CA. Restoration of threatened species: a noble cause for transgenic trees. Tree Genet Genomes. 2007;3:111–8.
Miranda-Fontaina ME, Fernández-López J, Vettraino AM, Vannini A. Resistance of Castanea Clones to Phytophthora cinnamomi: testing and genetic control. Silvae Genet. 2007;56:11.
Cuenca B, Ocaña L, Salinero C, Pintos C, Mansilla JP, Rial C. Selection of Castanea sativa Mill. for resistance to Phytophthora cinnamomi: Micropropagation and testing of selected clones. In: ISHS Acta Hortic. Leuven, Belgium: 866 I Eur. Congr. Chestnut - Castanea 2009; 2010. p. 111–9.
Shearer BL, Crane CE, Cochrane JA, Dunne CP. Variation in susceptibility of threatened flora to Phytophthora cinnamomi. Australas Plant Pathol Springer Netherlands. 2013;42:491–502.
Frampton J, Isik F, Benson DM. Genetic variation in resistance to Phytophthora cinnamomi in seedlings of two Turkish Abies species. Tree Genet Genomes. 2013;9:53–63.
Dunn CP. The elms: breeding, conservation, and disease management. Springer US. 2000.
Nielsen LR, Kjær ED. Gene flow and mating patterns in individuals of wych elm (Ulmus glabra) in forest and open land after the influence of Dutch elm disease. Conserv Genet. 2010;11:257–68.
Brasier CM. Rapid evolution of introduced plant pathogens via interspecific hybridization is leading to rapid evolution of Dutch elm disease and other fungal plant pathogens. Bioscience. 2001;51:123–33.
Mittempergher L, Santini A. The history of elm breeding. For Syst. 2004;13:161–77.
Heybroek H. The Dutch elm breeding program. In: Sticklen M, Sherald J, editors. Dutch Elm Dis. Res. US: Springer; 1993. p. 16–25.
Collin E, Bozzano M. Implementing the dynamic conservation of elm genetic resources in Europe: case studies and perspectives. iForest - Biogeosciences For. 2015;8:143–8.
McLachlan JS, Hellmann JJ, Schwartz MW. A framework for debate of assisted migration in an era of climate change. Conserv Biol. 2007;21:297–302.
Martín JA, Solla A, Venturas M, Collada C, Domínguez J, Miranda E, et al. Seven Ulmus minor clones tolerant to Ophiostoma novo-ulmi registered as forest reproductive material in Spain. iForest - Biogeosciences For. 2015;8:172–80.
Venturas M, López R, Martín JA, Gascó A, Gil L. Heritability of Ulmus minor resistance to Dutch elm disease and its relationship to vessel size, but not to xylem vulnerability to drought. Plant Pathol. 2014;63:500–9.
Townsend AM, Bentz SE, Douglass LW. Evaluation of 19 American elm clones for tolerance to Dutch elm disease. Enviornmental Hortic. 2005;23:21–4.
Siegert NW, McCullough DG, Liebhold AM, Telewski FW. Resurrected from the ashes: a historical reconstruction of emerald ash borer dynamics through dendrochronological analysis. Emerald ash borer Asian longhorned beetle Res. Dev. Rev. Meet. Cincinnatti, OH. FHTET 2007–04. Morgantown, WV U.S.: For. Serv. For. Heal. Technol. Enterp. Team; 2007. p. 18–9.
Haack R, Jendek E, Liu H, Marchant K. The emerald ash borer: a new exotic pest in North America. Newsl Michigan Entomol Soc. 2002;47:1–5.
Xia W, Reardon D, Wu Y, JiangHua S. Emerald ash borer, Agrilus planipennis Fairmaire (Coleoptera: Buprestidae) in China: a review and distribution survey. Acta Entomol Sin. 2004;47:679–85.
Poland TM, McCullough DG. Emerald ash borer: invasion of the urban forest and the threat to North America’s ash resource. J For. 2006;104:118–24.
Anulewicz AC, McCullough DG, Cappaert DL. Emerald ash borer (Agrilus planipennis) density and canopy dieback in three North American ash species. Arboric Urban For. 2007;33:338–49.
Klooster WS, Herms DA, Knight KS, Herms CP, McCullough DG, Smith A, et al. Ash (Fraxinus spp.) mortality, regeneration, and seed bank dynamics in mixed hardwood forests following invasion by emerald ash borer (Agrilus planipennis). Biol Invasions. 2014;16:859–73.
Rebek EJ, Herms DA, Smitley DR. Interspecific variation in resistance to emerald ash borer (Coleoptera: Buprestidae) among North American and Asian ash (Fraxinus spp.). Environ Entomol. 2008;37:242–6.
Anulewicz AC, McCullough DG, Cappaert DL, Poland TM. Host range of the emerald ash borer (Agrilus planipennis Fairmaire) (Coleoptera: Buprestidae) in North America: results of multiple-choice field experiments. Environ Entomol. 2008;37:230–41.
Tanis SR, McCullough DG. Host resistance of five Fraxinus species to Agrilus planipennis (Coleoptera: Buprestidae) and effects of Paclobutrazol and fertilization. Environ Entomol. 2015;44:287–99.
Koch JL, Mason ME, Carey DW, Knight K, Poland T, Herms DA, et al. Survey for tolerance to emerald ash borer within North American ash species. Proc. Symp. ash North Am In: Gen. Tech. Rep., Northern Research Station, US. 2010. p. 60.
Koch JL, Carey DW, Mason ME, Poland TM, Knight KS. Intraspecific variation in Fraxinus pennsylvanica responses to emerald ash borer (Agrilus planipennis). New For. Springer Netherlands; 2015; doi: 10.1007/s11056-015-9494-4
Baranchikov Y, Mozolevskaya E, Yurchenko G, Kenis M. Occurrence of the emerald ash borer, Agrilus planipennis in Russia and its potential impact on European forestry. EPPO Bull. 2008;38:233–8.
Straw NA, Williams DT, Kulinich O, Gninenko YI. Distribution, impact and rate of spread of emerald ash borer Agrilus planipennis (Coleoptera: Buprestidae) in the Moscow region of Russia. Forestry. 2013;86:515–22.
Havill NP, Montgomery ME, Yu G, Shiyake S, Caccone A. Mitochondrial DNA from Hemlock woolly adelgid (Hemiptera: Adelgidae) suggests cryptic speciation and pinpoints the source of the introduction to Eastern North America. Ann Entomol Soc Am. 2006;99:195–203.
Spaulding HL, Rieske LK. The aftermath of an invasion: structure and composition of Central Appalachian hemlock forests following establishment of the hemlock woolly adelgid. Adelges tsugae Biol Invasions. 2010;12:3135–43.
Domec JC, Rivera LN, King JS, Peszlen I, Hain F, Smith B, et al. Hemlock woolly adelgid (Adelges tsugae) infestation affects water and carbon relations of eastern hemlock (Tsuga canadensis) and Carolina hemlock (Tsuga caroliniana). New Phytol. 2013;199:452–63.
Orwig D, Foster D. Forest response to the introduced hemlock woolly adelgid in southern New England, USA. J Torrey Bot Soc. 1998;125:60–73.
Orwig D, Foster D. Stand, landscape, and ecosystem analyses of hemlock woolly adelgid outbreaks in southern New England: an overview. Proc Symp Sustain Manag Hemlock Ecosyst East North Am. 2000;267:123–5.
Griffin S. Hemlock Woolly Adelgids in Georgia. Georgia forestry commission. 2007. Online: http://www.gfc.state.ga.us/forest-management/forest-health/hemlock-woolly-adelgid/HWAinGeorgiaFactsheet07r.pdf
Cheah C, Montgomery M, Salem S, Parker B, Skinner M, Costa S. Biological control of hemlock woolly adelgid. For. Heal. Technol. Enterp. Team. 2004. 1–19
Orwig DA, Foster DR, Mausel DL. Landscape patterns of hemlock decline in New England due to the introduced hemlock woolly adelgid. J Biogeogr. 2002;29:1475–87.
Stadler B, Müller T, Orwig D, Cobb R. Hemlock woolly adelgid in New England forests: canopy impacts transforming ecosystem processes and landscapes. Ecosystems. 2005;8:233–47.
Mcclure MS. Reshaping the ecology of invading populations of hemlock woolly adelgid, Adelges tsugae (Homoptera: Adelgidae), in eastern North America. Biol. Invasions. 1999; 247–54
Caswell T, Casagrande R, Maynard B, Preisser E. Production and evaluation of eastern hemlocks potentially resistant to the hemlock woolly adelgid. United States, Hartford: 4th Symp. hemlock woolly adelgid East; 2008. p. 124–34.
Ingwell L, Preisser E. Using citizen science programs to identify host resistance in pest‐invaded forests. Conserv Biol Blackwell Publishing Inc. 2011;25:182–8.
Pontius JA, Hallett RA, Jenkins JC. Foliar chemistry linked to infestation and susceptibility to hemlock woolly adelgid (Homoptera: Adelgidae). Environ Entomol. 2006;35:112–20.
McKenzie EA, Elkinton JS, Casagrande RA, Preisser EL, Mayer M. Terpene chemistry of eastern hemlocks resistant to hemlock woolly adelgid. J Chem Ecol. 2014;40:1003–12.
Dukes JS, Pontius J, Orwig D, Garnas JR, Rodgers VL, Brazee N, et al. Responses of insect pests, pathogens, and invasive plant species to climate change in the forests of northeastern North America: what can we predict? This article is one of a selection of papers from NE Forests 2100: A Synthesis of Climate Change Impacts o. Can J For Res. 2009;39:231–48.
Mamiya Y. Pathology of the pine wilt disease caused by Bursaphelenchus xylophilus. Annu Rev Phytopathol. 1983;21:201–20.
Zhao T, Krokene P, Hu J, Christiansen E, Björklund N, Långström B, et al. Induced terpene accumulation in Norway spruce inhibits bark beetle colonization in a dose-dependent manner. PLoS One. 2011;6:e26649.
Burgermeister W, Sousa E, Mota M, Penas AC, Bravo MA, Metge K, et al. First report of Bursaphelenchus xylophilus in Portugal and in Europe. Nematology. 1999;1:727–34.
Rodrigues J. National eradication programme for the Pinewood nematode. In: Pine wilt disease—A worldwide threat to forest ecosystems. Springer Netherlands. 2008. p. 5–14
Vicente C, Espada M, Vieira P, Mota M. Pine wilt disease: a threat to European forestry. Eur J Plant Pathol. 2012;133:89–99.
Sousa E, Bravo M, Pires J, Naves P, Penas A, Bonifácio L, et al. Bursaphelenchus xylophilus (Nematoda; Aphelenchoididae) associated with Monochamus galloprovincialis (Coleoptera; Cerambycidae) in Portugal. Nematology. 2001;3:89–91.
Shi J, Luo Y, Xia N, Wu H, Song J. Suggestions on management measures of pine forest ecosystems invaded by Bursaphelenchus xylophilus. For Stud China. 2008;10:45–8.
Nose M, Shiraishi S. Comparison of the gene expression profiles of resistant and non-resistant Japanese black pine inoculated with pine wood nematode using a modified LongSAGE technique. For Pathol. 2011;41:143–55.
Zhang F, Kajiwara J, Mori Y, Ohira M, Tsutsumi Y, Kondo R. Metabolites from resistant and susceptible Pinus thunbergii after inoculation with pine wood nematode. Am J Plant Sci. 2013;4:512–8.
Nose M, Shiraishi S. Breeding for resistance to pine wilt disease. In: Pine wilt disease. Springer Japan. 2008. p. 334–50.
Da Silva M, Solla A, Sampedro L, Zas R, Vasconcelos MW. Susceptibility to the pinewood nematode (PWN) of four pine species involved in potential range expansion across Europe. Tree Physiol. 2015;35:987–99.
Zas R, Moreira X, Ramos M, Lima MRM, Nunes da Silva M, Solla A, et al. Intraspecific variation of anatomical and chemical defensive traits in Maritime pine (Pinus pinaster) as factors in susceptibility to the pinewood nematode (Bursaphelenchus xylophilus). Trees. 2015;29:663–73.
Rutherford TA, Webster JM. Distribution of pine wilt disease with respect to temperature in North America, Japan, and Europe. Can J For Res. 1987;17:1050–9.
Evans H, McNamara D, Braasch H, Chadoeuf J, Magnusson C. Pest risk analysis (PRA) for the territories of the European Union (as PRA area) on Bursaphelenchus xylophilus and its vectors in the genus Monochamus. Bull OEPP/EPPO. 1996;26:199–249.
Santini A, Fagnani A, Ferrini F, Ghelardini L, Mittempergher L. Variation among Italian and French elm clones in their response to Ophiostoma novo-ulmi inoculation. For Pathol. 2005;35:183–93.
Ghelardini L, Santini A. Avoidance by early flushing: a new perspective on Dutch elm disease research. IForest. 2009;2:143–53.
Hamilton M. Chapter 3. Genetic drift and effective population size. Popul. Genet. Wiley & Blackwell; 2009. p. 424.
Dodd RS, Hüberli D, Mayer W, Harnik TY, Afzal-Rafii Z, Garbelotto M. Evidence for the role of synchronicity between host phenology and pathogen activity in the distribution of sudden oak death canker disease. New Phytol. 2008;179:505–14.
Parker A, Goudie AS, Anderson DE, Robinson MA, Bonsall C. A review of the mid-Holocene elm decline in the British Isles. Prog Phys Geogr. 2002;26:1–45.
Rajala T, Velmala SM, Vesala R, Smolander A, Pennanen T. The community of needle endophytes reflects the current physiological state of Norway spruce. Fungal Biol. 2014;118:309–15.
Gennaro M, Gonthier P, Nicolotti G. Fungal endophytic communities in healthy and declining Quercus robur L. and Q. cerris L. trees in Northern Italy. J Phytopathol. 2003;151:529–34.
Martín JA, Witzell J, Blumenstein K, Rozpedowska E, Helander M, Sieber TN, et al. Resistance to Dutch elm disease reduces presence of xylem endophytic fungi in Elms (Ulmus spp.). PLoS One. 2013;8:e56987.
Díaz G, Córcoles AI, Asencio AD, Torres MP. In vitro antagonism of Trichoderma and naturally occurring fungi from elms against Ophiostoma novo-ulmi. For Pathol. 2013;43:51–8.
Witzell J, Martín JA, Blumenstein K. Ecological aspects of endophyte-based biocontrol of forest diseases. In: Advances in Endophytic Research. 2014. Springer India. p. 321–33. This book chapter provides insights on the role of endophytes in tree resistance to pests and pathogens and highlights the urgent need for research on endophyte-based biocontrol of forest tree diseases.
IUFRO. Montesclaros Declaration. 2011, http://www.iufro.org/science/divisions/division-7/70000/publications/montesclaros-declaration/
Wingfield MJ, Brockerhoff EG, Wingfield BD, Slippers B. Planted forest health: the need for a global strategy. Science. 2015;349:832–6.
Nielsen LR, Kjær ED. Fine-scale gene flow and genetic structure in a relic Ulmus laevis population at its northern range. Tree Genet Genomes. 2010;6:643–9.
Bacles CFE, Ennos RA. Paternity analysis of pollen-mediated gene flow for Fraxinus excelsior L. in a chronically fragmented landscape. Heredity. 2008;101:368–80.
Lowe AJ, Cavers S, Boshier D, Breed MF, Hollingsworth PM. The resilience of forest fragmentation genetics—no longer a paradox—we were just looking in the wrong place. Heredity. 2015;115:97–9.
Cavers S. Evolution, ecology and tree health: finding ways to prepare Britain’s forests for future threats. Forestry. 2015;88:1–2.
Gil L, Fuentes-Utrillo P, Soto A, Cervera MT, Collada C. Phylogeography: English elm is a 2,000-year-old Roman clone. Nature. 2004;431:1053.
Alexander J, Lee CA. Lessons learned from a decade of sudden oak death in California: evaluating local management. Environ Manage. 2010;46:315–28.
Meentemeyer RK, Dorning MA, Vogler JB, Schmidt D, Garbelotto M. Citizen science helps predict risk of emerging infectious disease. Front Ecol Environ. 2015;13:189–94.
Haack RA, Hérard F, Sun J, Turgeon JJ. Managing invasive populations of Asian longhorned beetle and citrus long-horned beetle: a worldwide perspective. Annu Rev Entomol. 2010;55:521–46.
Schröder T. Zur aktuellen Situation des Kiefernholznematoden Bursaphelenchus xylophilus in der EU- Sachstand, Risikobewertung und Managementoptionen. Julius-Kuhn-Archiv. 2010;428:234.
Vannatta AR, Hauer RH, Schuettpelz NM. Economic analysis of emerald ash borer (Coleoptera: Buprestidae) management options. J Econ Entomol. 2012;105:196–206.
Foster D, Orwig D. Preemptive and salvage harvesting of New England forests: when doing nothing is a viable alternative. Conserv Biol. 2006;20:959–70.
Havill N, Montgomery M. The role of Arboreta in studying the evolution of host resistance to the hemlock woolly adelgid. Arnoldia. 2008;65:2–9.
COST action FP1401 “A global network of nurseries as early warning system against alien tree pests (Global warning)”, webpage: http://www.cost.eu/COST_Actions/fps/FP1401
REINFFORCE - REsource INFrastructure for monitoring and adapting European Atlantic FORests under Changing climate, webpage: http://www.forestry.gov.uk/fr/reinfforce
Drenkhan R, Hanso M. New host species for Chalara fraxinea. New Dis Reports. 2010;22:16.
Koskela J, Lefèvre F, Schueler S, Kraigher H, Olrik DC, Hubert J, et al. Translating conservation genetics into management: Pan-European minimum requirements for dynamic conservation units of forest tree genetic diversity. Biol Conserv. 2013;157:39–49.
Graudal LOV, Kjær ED, Thomsen A, Larsen B. Planning national programmes for conservation of forest genetic resources. Humlebæk, Denmark: Danida For. Seed Centre, Tech. notes 46. Danida Forest Seed Centre; 1997.
Frankham R, Bradshaw CJA, Brook BW. Genetics in conservation management: revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol Conserv. 2014;170:56–63.
Graudal L, Kjær ED, Canger S. A systematic approach to the conservation of genetic resources of trees and shrubs in Denmark. Forest Ecol Manag. 1995;73:117–34.
Kjær E, Amaral W, Yanchuk A, Graudal L. Strategies for conservation of forest genetic resources. For Genet Resour Conserv Manag. 2004;1:5–24. FAO, Rome.
Acknowledgments
Chatchai Kosawang is thanked for his comments to the manuscript regarding the role of endophytes. This work was supported by the Villum foundation in the framework of the ‘Trees for the future forests’ project [VKR023062]. We also thank two anonymous reviewers and the editor for helpful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors report grants from Villum foundation during the course of this review.
Human and Animal Rights and Informed Consent
The article contains no studies with human or animal subjects performed by the authors.
Additional information
This article is part of the Topical Collection on Tree Genetics
Rights and permissions
About this article
Cite this article
Budde, K.B., Nielsen, L.R., Ravn, H.P. et al. The Natural Evolutionary Potential of Tree Populations to Cope with Newly Introduced Pests and Pathogens—Lessons Learned From Forest Health Catastrophes in Recent Decades. Curr Forestry Rep 2, 18–29 (2016). https://doi.org/10.1007/s40725-016-0029-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40725-016-0029-9