Abstract
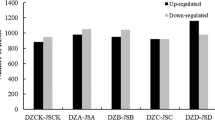
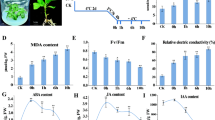
Freezing stress is a major factor affecting plant growth, crop productivity, and the geographical distribution of plants. To identify freezing-responsive genes in Brassica rapa, we analyzed transcriptome profiles of two contrasting inbred lines with different geographic origins, Chiifu and Kenshin, in control and −4 °C-treated leaves. A total of 3,301 genes were differentially expressed between Chiifu and Kenshin upon freezing treatment. Among these, 67 and 1,633 genes were specifically expressed in Chiifu and Kenshin, respectively. An ortholog (BrTPP1) of Arabidopsis trehalose-6-phosphate phosphatase 1 (TPP1) was specifically and highly induced in Chiifu by freezing treatment. However, most cold-responsive genes, including CBF pathway-related genes, showed similar patterns of expression between Chiifu and Kenshin. Many genes involved in stress responses (i.e., to freezing temperatures) were intrinsically and specifically expressed in Chiifu, which is tolerant of freezing temperatures. The results suggest that the CBF pathway is not the main factor conferring freezing tolerance to Chiifu, but genes that are expressed prior to cold acclimation, along with other regulatory genes, may play important roles in freezing tolerance.




Similar content being viewed by others
References
Anchorodoguy TJ, Rudolph AS, Carpenter JF, Crowe JH (1987) Modes of interaction of cryoprotectants with membrane phospholipids during freezing. Cryobiology 24:324–331
Bae MS, Cho EJ, Choi EY, Park OK (2003) Analysis of the Arabidopsis nuclear proteome and its response to cold stress. Plant J 36:652–663
Bita CE, Zenoni S, Vriezen WH, Mariani C, Pezzotti M, Gerats T (2011) Temperature stress differentially modulates transcription in meiotic anthers of heat-tolerant and heat-sensitive tomator plants. BMC Genomics 12:384
Browse J, Xin Z (2001) Temperature sensing and cold acclimation. Curr Opin Plant Biol 4:241–246
Capel J, Jarillo JA, Salinas J, Martinez-Zapater JM (1997) Two homologous low-temperature-inducible genes from Arabidopsis encode highly hydrophobic proteins. Plant Physiol 115:569–576
Carvallo MA, Pino MT, Jeknic Z, Zou C, Doherty CJ, Shiu SH, Chen THH, Thomashow MF (2011) A comparison of the low temperature transcriptomes and CBF regulons of three plant species that differ in freezing tolerance: Solanum commersonii, Solanum tuberosum, and Arabidopsis thaliana. J Exp Bot 62:3807–3819
Cavender-Bares J (2007) Chilling and freezing stress in live oaks (Quercus section Virentes): intra- and inter-specific variation in PSII sensitivity corresponds to latitude of origin. Photosynth Res 94:437–453
Chen CN, Chu CC, Zentella R, Pan SM, Ho THD (2002) AtHVA22 gene family in Arabidopsis: phylogenetic relationship, ABA and stress regulation, and tissue-specific expression. Plant Mol Biol 49:633–644
Chen QF, Xiao S, Chye ML (2008) Overexpression of the Arabidopsis 10-kilodalton acyl-coenzyme A-binding protein ACBP6 enhances freezing tolerance. Plant Physiol 148:304–315
Chiba Y, Mineta K, Hirai MY, Suzuki Y, Kanaya S, Takahashi H, Onouchi H, Yamaguchi J, Naito S (2013) Changes in mRNA stability associated with cold stress in Arabidopsis cells. Plant Cell Physiol 54:180–194
Chinnusamy V, Zhu JK, Sunkar R (2010) Gene regulation during cold stress acclimation in plants. Methods Mol Biol 639:39–55
Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14:1675–1690
Frank G, Pressman E, Ophir R, Althan L, Shaked R, Freedman M, Shen S, Firon N (2009) Transcriptional profiling of maturing tomato (Solanum lycopersicum L.) microspores reveals the involvement of heat shock proteins, ROS scavengers, hormones, and sugars in the heat stress response. J Exp Bot 60:3891–3908
Garg AK, Kim JK, Owens TG, Ranwala AP, Choi YD, Kochian LV, Wu RJ (2002) Trehalose accumulation in irce plants confers high tolerance levels to different abiotic stresses. Proc Natl Acad Sci USA 99:15898–15903
Goulas E, Schubert M, Kieselbach T, Kleczkowski LA, Gardestrom P, Schroder W, Hurry V (2006) The chloroplast lumen and stromal proteomes of Arabidopsis thaliana show differential sensitivity to short- and long-term exposure to low temperature. Plant J 47:720–734
Iordachescu M, Imai R (2008) Trehalose biosynthesis in response to abiotic stresses. J Inetgr Plant Biol 50:1223–1229
Jang IC, Oh SJ, Seo JS, Choi WB, Song SI, Kim CH, Kim YS, Seo HS, Choi YD, Nahm BH, Kim JK (2003) Expression of a bifunctional fusion of the Escherichia coli genes for trehalose-6-phosphate synthase and tranhalose-6-phosphate phosphatase in transgenic rice plants increases trehalose accumulation and abiotic stress tolerance withoud stunting growth. Plant Physiol 131:516–524
Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, Guy CL (2004) Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol 136:4159–4168
Karlson D, Imai R (2003) Conservation of the cold shock domain protein family in plants. Plant Physiol 131:12–15
Kawamura Y, Uemura M (2003) Mass spectrometric approach for identifying putative plasma membrane proteins of Arabidopsis leaves associated with cold acclimation. Plant J 36:141–154
Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50:347–363
Kodama H, Hamada T, Horiguchi G, Nishimura M, Iba K (1994) Genetic enhancement of cold tolerance by expression of a gene for chloroplast omega-3-fatty-acid desaturase in transgenic tobacco. Plant Physiol 105:601–605
Kumari S, Panjabi V, Kushwaha H, Sopory S, Singla-Pareek S, Pareek A (2009) Transcriptome map for seedling stage specific salinity stress response indicates a specific set of genes as candidate for saline tolerance in Oryza sativa L. Funct Integr Genomic 9:109–123
Lee H, Guo Y, Ohta M, Xiong LM, Stevenson B, Zhu JK (2002) LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase. EMBO J 21:2692–2702
Lee BH, Henderson DA, Zhu JK (2005) The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17:3155–3175
Lee SC, Lim MH, Kim JA, Lee SI, Kim JS, Jin M, Kwon SJ, Mun JH, Kim YK, Kim HU, Hur Y, Park BS (2008) Transcriptome analysis in Brassica rapa under abiotic stresses using Brassica 24K oligo microarray. Mol Cells 26:595–605
Lee J, Lim YP, Han CT, Nou IS, Hur Y (2013) Genome-wide expression profiles of contransting inbred lines of Chinese cabbage, Chiifu and Kenshin, under temperature stress. Genes Genom 35:265–281
Lissarre M, Ohta M, Sato A, Miura K (2010) Cold-responsive gene regulation during cold acclimation in plants. Plant Signal Behav 5:948–952
Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, Shimada Y, Yoshida S, Shinozaki K, Yamaguchi-Shinozaki K (2004) Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J 38:982–993
Medina J, Catalá R, Salinas J (2011) The CBFs: three arabidopsis transcription factors to cold acclimate. Plant Sci 180:3–11
Miranda JA, Avonce N, Suárez R, Thevelein JM, Van Dijck P, Iturriaga G (2007) A biosunctional TPS-TPP enzyme from yeast confers tolerance to multiple and extreme abiotic-stress conditions in transgenic Arabidopsis. Planta 226:1411–1421
Mizoguchi T, Irie K, Hirayama T, Hayashida N, Yamaguchi-Shinozaki K, Matsumoto K, Shinozaki K (1996) A gene encoding a mitogen-activated protein kinase kinase kinase is induced simultaneously with genes for a mitogen-activated protein kinase and an S6 ribosomal protein kinase by touch, cold, and water stress in Arabidopsis thaliana. Proc Natl Acad Sci USA 93:765–769
Moellering ER, Muthan B, Benning C (2010) Freezing tolerance in plants requires lipid remodeling at the outer chloroplast membrane. Science 330:226–228
Monroy AF, Dryanova A, Malette B, Oren DH, Ridha Farajalla M, Liu W, Danyluk J, Ubayasena LW, Kane K, Scoles GJ, Sarhan F, Gulick PJ (2007) Regulatory gene candidates and gene expression analysis of cold acclimation in winter and spring wheat. Plant Mol Biol 64:409–423
Nishizawa A, Yabuta Y, Shigeoka S (2008) Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol 147:1251–1263
Oono Y, Seki M, Satou M, Iida K, Akiyama K, Sakurai T, Fujita M, Yamaguchi-Shinozaki K, Shinozaki K (2006) Monitoring expression profiles of Arabidopsis genes during cold acclimation and deacclimation using DNA microarrays. Funct Intergr Genomics 6:212–234
Partridge M, Murphy DJ (2009) Roles of a membrane-bound caleosin and putative peroxygenase in biotic and abiotic stress responses in Arabidopsis. Plant Physiol Biochem 47:796–806
Robinson SJ, Parkin IA (2008) Differential SAGE analysis in Arabidopsis uncovers increased transcriptome complexity in response to low temperature. BMC Genomics 9:434
Sanghera GS, Wani SH, Hussain W, Singh NB (2011) Engineering cold stress tolerance in crop plants. Curr Genomics 12:30–43
Sharma P, Sharma N, Deswal R (2005) The molecular biology of the low-temperature response in plants. BioEssays 27:1048–1059
Sharma N, Cram D, Huebert T, Zhou N, Parkin IA (2007) Exploiting the wild crucifer Thlaspi arvense to identify conserved and novel genes expressed during a plant’s response to cold stress. Plant Mol Biol 63:171–184
Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3:217–223
Taji T, Seki M, Satou M, Sakurai T, Kobayashi M, Ishiyama K, Narusaka Y, Narusaka M, Zhu JK, Shinozaki K (2004) Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol 135:1697–1709
Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol 50:571–599
Thomashow MF (2010) Molecular basis of plant cold climation: insights gained from studying the CBF cold response pathway. Plant Physiol 154:571–577
Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41:195–211
Winfield MO, Lu C, Wilson ID, Coghill JA, Edwards KJ (2010) Plant responses to cold: transcriptome analysis of wheat. Plant Biotechnol J 8:749–771
Xin Z, Browse J (1998) eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc Natl Acad Sci USA 95:7799–7804
Xin Z, Browse J (2000) Cold comfort farm: the acclimation of plants to freezing temperatures. Plant Cell Environ 23:893–902
Zhang X, Byrnes JK, Gal TS, Li WH, Borevitz JO (2008) Whole genome transcriptome polymorphisms in Arabidopsis thaliana. Genome Biol 9(11):R165
Zhang T, Zhao XQ, Wang WS, Pan YJ, Huang LY, Liu XY, Zong Y, Zhu LH, Yang DC, Fu BY (2012) Comparative transcriptome profiling of chilling atress responsiveness in two xontrasting rice fenotypes. PLoS ONE 7(8):e43274
Zhen Y, Ungerer MC (2008) Clinal variation in freezing tolerance among natural accessions of Arabidopsis thaliana. New Phytol 177:419–427
Zhou MQ, Shen C, Wu LH, Tang KX, Lin J (2011) CBF-dependent signaling pathway: a key responder to low temperature stress in plants. Crit Rev Biotechnol 31:186–192
Zhu J, Dong CH, Zhu JK (2007) Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr Opin Plant Biol 10:290–295
Zuther E, Schulz E, Childs LH, Hincha DK (2012) Clinal variation in the non-acclimated and cold-acclimated freezing tolerance of Arabidopsis thaliana accessions. Plant Cell Environ 35:1860–1878
Acknowledgments
This work was supported by a Grant from the Next-Generation BioGreen 21 Program (the Next-Generation Genomics Center no. PJ008118 and Plant Molecular Breeding Center no. PJ009085), Rural Development Administration, Republic of Korea.
Conflict of interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dong, X., Im, SB., Lim, YP. et al. Comparative transcriptome profiling of freezing stress responsiveness in two contrasting Chinese cabbage genotypes, Chiifu and Kenshin. Genes Genom 36, 215–227 (2014). https://doi.org/10.1007/s13258-013-0160-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-013-0160-y