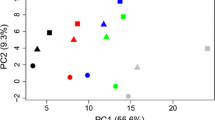
Abstract
Freezing tolerance in plants develops through acclimation to cold by growth at low, above-freezing temperatures. Wheat is one of the most freezing-tolerant plants among major crop species and the wide range of freezing tolerance among wheat cultivars makes it an excellent model for investigation of the genetic basis of cold tolerance. Large numbers of genes are known to have altered levels of expression during the period of cold acclimation and there is keen interest in deciphering the signaling and regulatory pathways that control the changes in gene expression associated with acquired freezing tolerance. A 5740 feature cDNA amplicon microarray that was enriched for signal transduction and regulatory genes was constructed to compare changes in gene expression in a highly cold-tolerant winter wheat cultivar CDC Clair and a less tolerant spring cultivar, Quantum. Changes in gene expression over a time course of 14 days detected over 450 genes that were regulated by cold treatment and were differentially regulated between spring and winter cultivars, of these 130 are signaling or regulatory gene candidates, including: transcription factors, protein kinases, ubiquitin ligases and GTP, RNA and calcium binding proteins. Dynamic changes in transcript levels were seen at all periods of cold acclimation in both cultivars. There was an initial burst of gene activity detectable during the first day of CA, during which 90% of all genes with increases in transcript levels became clearly detectable and early expression differential between the two cultivars became more disparate with each successive period of cold acclimation.






Similar content being viewed by others
References
Bottley A, Xia GM, Koebner RM (2006) Homoeologous gene silencing in hexaploid wheat. Plant J 47:897–906
Brown RS (2005) Zinc finger proteins: getting a grip on RNA. Curr Opin Struct Biol 15:94–98
Chinnusamy V, Ohta M, Kanrar S et al (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17:1043–1054
Cook D, Fowler S, Fiehn O et al (2004) A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc Natl Acad Sci USA 101:15243–15248
Danyluk J, Kane NA, Breton G et al (2003) TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol 132:1849–1860
de Folter S, Immink RG, Kieffer M et al (2005) Comprehensive Interaction Map of the Arabidopsis MADS Box Transcription Factors. Plant Cell 17:1424–1433
Eisen MB, Spellman PT, Brown PO et al (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95:14863–14868
Faris JD, Haen KM, Gill BS (2000) Saturation mapping of a gene-rich recombination hot spot region in wheat. Genetics 154:823–835
Fowler DB, Limin AE, Shi-Ying Wang S-Y et al (1996) Relationship between low-temperature tolerance and vernalization response in wheat and rye. Can J Plant Sci 76:37–42
Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14:1675–1690
Fu D, Szucs P, Yan L et al (2005) Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Genet Genomics 273:54–65
Griffith M, Lumb C, Wiseman SB, Wisniewski M, Johnson RW, Marangoni AG (2005) Antifreeze proteins modify the freezing process in planta. Plant Physiol 138:330–340
Gulick PJ, Drouin S, Yu Z et al (2005) Transcriptome comparison of winter and spring wheat responding to low temperature. Genome 48:913–923
Hellmann H, Estelle M (2002) Plant development: regulation by protein degradation. Science 297:793–797
Limin AE, Fowler DB (1985) Cold hardiness in Triticum and Agelops species. Can J Plant Sci 65:71–78
Martin ML, Busconi L (2001) A rice membrane-bound calcium-dependent protein kinase is activated in response to low temperature. Plant Physiol 125:1442–1449
Monroy A, Dhindsa RS (1995) Low-temperature signal transduction: induction of cold acclimation-specific genes of alfalfa by calcium at 25°C. Plant Cell 7:321–331
Olsen AN, Ernst HA, Leggio LL et al (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10:79–87
Oono Y, Seki M, Satou M et al (2006) Monitoring expression profiles of Arabidopsis genes during cold acclimation and deacclimation using DNA microarrays. Funct Integr Genomics 6:212–234
Prasil IT, Prasilova P, Pankova K (2004) Relationships among vernalization, shoot apex development and frost tolerance in wheat. Ann Bot 94:413–418
Qi LL, Echalier B, Chao S et al (2004) A chromosome bin map of 16,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics 168:701–712
Riechmann JL, Ratcliffe OJ (2000) A genomic perspective on plant transcription factors. Curr Opin Plant Biol 3:423–434
Saijo Y, Hata S, Kyozuka J et al (2000) Over-expression of a single Ca2_-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23:319–327
Seki M, Narusaka M, Ishida J et al 2002 Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31:279–292
Shin D, Koo YD, Lee J et al (2004) Athb-12, a homeobox-leucine zipper domain protein from Arabidopsis thaliana, increases salt tolerance in yeast by regulating sodium exclusion. Biochem Biophys Res Commun 323:534–540
Shiu SH, Karlowski WM, Pan R et al (2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16:1220–1234
Shou H, Bordallo P, Fan JB et al (2004) Expression of an active tobacco mitogen-activated protein kinase kinase kinase enhances freezing tolerance in transgenic maize. Proc Natl Acad Sci USA 101:3298–3303
Simon RM, Dobbin K (2003) Experimental design of DNA microarray experiments. Biotechniques Mar Suppl:16–21
Simpson GG, Quesada V, Henderson IR et al (2004) RNA processing and Arabidopsis flowering time control. Biochem Soc Trans 32:565–566
Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100:9440–9445
Teige M, Scheikl E, Eulgem T et al (2004) The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell 15:141–152
Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50:571–599
Tran LS, Nakashima K, Sakuma Y et al (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16:2481–2498
Tremblay K, Ouellet F, Fournier J, Danyluk J, Sarhan F (2005) Molecular characterization and origin of novel bipartite cold-regulated ice recrystallization inhibition proteins from cereals. Plant Cell Physiol 46:884–891
Umemura Y, Ishiduka T, Yamamoto R et al (2004) The Dof domain, a zinc finger DNA-binding domain conserved only in higher plants, truly functions as a Cys2/Cys2 Zn finger domain. Plant J 37:741–749
Vannini C, Locatelli F, Bracale M et al (2004) Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J 37:115–127
Vogel JT, Zarka DG, van Buskirk HA et al (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41:195–211
Yan L, Loukoianov A, Blechl A et al (2004) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303:1640–1644
Yan L, Loukoianov A, Tranquilli G et al (2003) Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100:6263–6268
Yang T, Chaudhuri S, Yang L et al (2004) Calcium/calmodulin up-regulates a cytoplasmic receptor-like kinase in plants. J Biol Chem 279:42552–42559
Zhu J, Shi H, Lee BH et al (2004) An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway. Proc Natl Acad Sci USA 101:9873–9878
Zhu J, Verslues PE, Zheng X et al (2005) HOS10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proc Natl Acad Sci USA 102:9966–9971
Acknowledgements
We thank Matt Links, Luke McCarthy, and Bill Crosby for assistance with bioinformatics for wheat EST sequences; Olin Anderson for cDNA clones used in the microarray construction, and Ian Ferguson for advice for statistical analysis. We thank Youko Oono, Motoaki Seki and Kazuo Shinozaki for providing gene expression data for Arabidopsis. We thank the Centre for Structural Genomics, Concordia University, for assistance in the preparation and printing of the microarray.
This work was supported by a Genome Canada, Genome Prairie, and Genome Quebec grant to P.J.G., G.J.S. and F.S. and by grants from the Natural Sciences and Engineering Council of Canada to P.J.G and F.S.
Antonio F. Monroy and Ani Dryanova contributed equally to this work.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material
11103_2007_9161_MOESM5_ESM.xls
All Features with Significant Cultivar x Cold Treatment Interaction Effect and Two Fold Change in Expression (XLS 290 kb)
Rights and permissions
About this article
Cite this article
Monroy, A.F., Dryanova, A., Malette, B. et al. Regulatory gene candidates and gene expression analysis of cold acclimation in winter and spring wheat. Plant Mol Biol 64, 409–423 (2007). https://doi.org/10.1007/s11103-007-9161-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-007-9161-z