Abstract
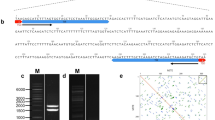
5S rRNA gene repeat units in a species are usually organized as either one relatively close size with numbers of intraspecific variations in NTS region or two different sizes with completely different sequence in NTS. Allium victorialis var. platyphyllum revealed two different size products of approximately 0.39 kb and 0.51 kb with highly conserved coding region of 120 bp. However, an extra sequences of approximately 120 bp between at 324 and 443 bp in long NTS region revealed, besides the remaining sequences of two NTS regions of short and long size were highly conserved giving the identity of 94.9%. To identify whether two different size 5S rDNA are occupied by a mixed state as random repeat or an independent group by each size in a particular locus, two rounds of FISH was sequentially performed using two probes of independent different size 5S rDNA and additional probe of only extra sequences of 120 bp in long NTS. Due to the highly conserved coding regions of both 5S rDNA, two different size 5S rDNA were detected in 3 loci in short arm of chromosome 6, however, extra sequences of long NTS was shown only in one locus within detected 5S rDNA from all examined chromosomes and interphase cells. This independent localization of two different size 5S rDNA suggests that 5S rDNA may be organized as a tandem repeat with random positions in a molecular level, but of cytogenetic view in chromosomes and interphase cells, they are organized as an independent group in a significant loci consisting of own size by the patterns of nucleotide variations.
Similar content being viewed by others
References
Appels R, Gerlach WL, Dennis ES, Swift H and Peacock WJ (1980) Molecular and chromosomal organization of DNA sequences coding for the ribosomal RNAs in cereals. Chromosoma 78: 293–311.
Appels R, Reddy P, McIntyre CL, Moran LB, Frankel OH and Clarke BC (1989) The molecular-cytogenetic analysis of grasses and its applications to studying relationships among species of the Triticeae. Genome 31: 122–133.
Baum BR and Bailey LG (2001) The origin of A genome donor of wheats (Triticum: Poaveae)-a prospective based on the sequence variation of the 5S DNA gene units. Gene Resource Crop Evol. 51: 183–196.
Baum BR, Bailey LG., Belyayev A, Raskina O and Nevo E (2004) The utility of the nontranscribed spacer of 5S rDNA units grouped into classes assigned to haplomes — a test on cultivated wheat and wheat progenitors. Genome 47: 590–599.
Baum BR and Johnson DA (1999) The 5S rRNA gene in wall barley (Hordeum murinum L. sensu lato): Sequence variation among repeat units and relationship to the Y haplome in the genus Hordeum (Porceae: Triticeae). Genome 42: 852–866.
Campbell CS, Wojciechowski MF, Baldwin BG, Alice A and Donoghue MJ (1997) Persistent nuclear ribosomal DNA sequences polymorphism in the Amelanchier agamic complex (Rosaceae). Mole. Biol. of Evol. 14: 81–90.
Cronn RC, Zhao X, Paterson AH and Wendel JF (1996) Polymorphism and concerted evolution in a tandemly repeated gene family: 5S ribosomal DNA in diploid and allopolyploid cottons. J. Mole. Evol. 42: 685–705
Do GS and Seo BB (2000) Phylogenetic relationships among Allium subg. Rhizirideum species based on the molecular variation of 5S rRNA genes. Korean J. Biol. Sci. 4: 77–85.
Do GS, Seo BB, Ko JM, Lee SH, Park JH, Kim IS and Song SD (1999) Analysis of Somaclonal variation through tissue culture and chromosomal localization of rDNA sites by fluorescent in situ hybridization in wild Allium tuberosum and a regenerated variant. Plant Cell Tissue Organ Cult. 57: 113–119.
Fritsch RM and Friesen N (2002) Evolution, domestication and taxonomy. In Allium crop science: recent advances, Ravinovitch H.D. and Currah, L. CAB international, Wallingford, U.K, pp. 5–27.
Goldborough PB, Ellis TH and Cullis CA (1981) Organization of the 5S RNA genes in flax. Nucleic Acids Res. 9: 5895–5904.
Hori H and Osawa S (1986) Evolutionary change in 5S rRNA secondary structure and a phylogenetic tree of 352 5S rRNA species. Biosystems 19: 163–172.
Kellogg EA and Appels R (1995) Intraspecific and interspecific variation in 5S RNA genes are decoupled in diploid wheat relatives. Genetics 140: 325–343.
Kim SY, Choi HW, Koo DH, Lee WK, Lee JK and Bang JW (2006) Characterization of eight Rumex species in FISH (Fluorescence in situ hybridization) and 5S rDNA spacer sequences. Kor. J. Genet. 22: 295–303.
Klass M (1998) Applications and impact of molecular markers on evolutionary and diversity studies in the genus Allium. Plant Breed. 117: 297–308.
Lapitan NLV, Ganal MW and Tanksley SD (1989) Somatic chromosome karyotypes of tomato based on in situ hybridization of the TAGI satellite repeat. Genome 32: 992–998.
Lee SH, Do GS and Seo BB (1999) Chromosomal localization of 5S rRNA gene loci and the implifications for relationships within the Allium complex. Chromosome Res. 7: 89–93.
Lee SH, Ryu JA, Do GS, Seo BB, Park JH, Kim IS and Song SD (1998) Chromosome analysis by fluorescence in situ hybridization of callus-derived regenerants in Allium cyaneum R. Plant Cell Rep. 18: 209–213.
Lee SH and Seo BB (1997) Chromosomal localization of 5S and 18S–26S rRNA genes using fluorescence in situ hybridization in Allium wakegi. Kor. J. Genet. 19: 19–26.
Leitch IJ and Heslop-Harrison JS (1993) Physical mapping of four sites of 5S ribosomal DNA sequences and one site of the x-amylase 2 gene in barley (Hordeum vulgare). Genome 36: 517–523.
Maughan PJ, Kolano BA, Maluszynska J, Coles ND, Bonifacio A, Rojas J, Coleman CE, Stevens MR, Fairbanks DJ, Parkinson SE, et al. (2006) Molecular and cytological characterization of ribosomal RNA genes in Chenopodium quinoa and Chenopodium berlandieri. Genome 49: 825–839.
Mes THM, Fritsch RM, Pollner S and Bachmann SK (1999) Evolution of the chloroplast genome and polymorphic ITS regions in Allium subg. Melanocrommyum. Genome 42: 237–247.
Mukai Y, Endo TR and Grill BS (1991) Physical mapping of the18S-26S rRNA multigene family in common wheat. J. Hered. 81: 290–295.
Park YK, Park HC, Park CH and Kim NS (2000) Chromosomal localization and sequence variation of 5S rRNA gene in five Capsicum species. Mol. Cells 10: 18–24.
Reddy AD, Suh SB, Ghaffari R, Singh NJ, Kim DJ, Han JH and Kim KS (2003) Bioinformatics analysis of SARS proteins and molecular dynamics simulated streucture of an alpha-helix motif. Bull. Kor. Chem. Soc. 24: 899–900.
Ricroch A, Peffley EBM and Barker RJ (1992) Chromosomal location of rDNA in Allium: in situ hybridization using biotin- and fluorescein-labelled probe. Theor. Appl. Genet. 83: 413–418.
Rogers SO and Bendich AJ (1987) Ribosomal gene in plants, variability in copy number and in the intergenic spacer. Plant Mol. Biol. 9: 509–520.
Samson ML and Wegnez M (1984) The 5S ribosomal genes in Drosophila melanogaster species subgroup. Nucleotide sequence of a 5S unit from Drosophila simulance and Drosophila teissieri. Nucleic acid Res. 12: 1003–1014.
Seo JH, Lee SY and Seo BB (2007a) Genome Analysis Using Sequence Variation and Localization of Tandem Repeats of 5S rRNA Gene in Allium wakegi. Kor. J. Breed Sci. 39: 70–76.
Seo JH, Park JH and Seo BB (2007b) Sequence variation among tandem repeat units of 5S rRNA gene and phylogenetic relationship in four taxa of Dendranthema. Kor. J. Genet. 29: 211–218.
Specht T, Wolters J and Erdmann VA (1990) Compilation of 5S rRNA and 5S rRNA gene sequences. Nucleic Acids Res. 18: 2215–2230.
Thompson JD, Higgins DG and Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 16: 486–499.
Yang YW, Tseng PF, Tai PY and Chang CJ (1998) Phylogenetic positions of Raphanus in relation to Brassica species based on 5S rRNA spacer sequence data. Bot. Bull. Acad. Sin. 39: 153–160.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Seo, J.H., Seo, B.B. Independent chromosomal localization of two different size 5S rDNA of Allium victorialis var. platyphyllum by sequential fluorescence in situ hybridization in accordance with sequence polymorphism. Genes Genom 32, 129–135 (2010). https://doi.org/10.1007/s13258-009-0804-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-009-0804-0