Abstract
Background
Miniature inverted repeat transposable elements (MITEs) are a dynamic component responsible for genome evolution. Tourist MITEs are derived from and mobilized by elements from the harbinger superfamily.
Objective
In this study, a novel family of Tourist-like MITE was characterized in wild soybean species Glycine falcata. The new GftoMITE1 was initially discovered as an insertional polymorphism of the centromere-specific histone H3 (CenH3) gene in G. falcata.
Methods
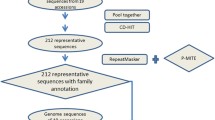
Using polymerase chain reaction, cloning and sequencing approaches, we showed a high number of copies of the GftoMITE1 family. Extensive bioinformatic analyses revealed the genome-level distribution and locus-specific mapping of GftoMITE1 members in Glycine species.
Results
Our results provide the first extensive characterization of the GftoMITE1 family and contribute to the understanding of the evolution of MITEs in the Glycine genus. Genome-specific GftoMITE1 was prominent in perennial wild soybean species, but not in annual cultivated soybean (Glycine max) or its progenitor (Glycine soja).
Conclusions
We discuss that the GftoMITE1 family reveals a single rapid amplification in G. falcata and could have potential implications for gene regulation and soybean breeding as an efficient genetic marker for germplasm utilization in the future.




Similar content being viewed by others
Data availability
All data are deposited in a publicly available repository with accession numbers.
References
Bureau TE, Wessler SR (1992) Tourist: A large family of small inverted repeat elements frequently associated with maize genes. Plant Cell 4(10):1283–1294. https://doi.org/10.1105/tpc.4.10.1283
Bureau TE, Wessler SR (1994) Stowaway: A new family of inverted repeat elements associated with the genes of both monocotyledonous and dicotyledonous plants. Plant Cell 6(6):907–916. https://doi.org/10.1105/tpc.6.6.907
Chao J-T, Kong Y-Z, Wang Q, Sun Y-H, Gong D-P, Lv J, Liu G-S (2015) MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Yi Chuan = Hereditas, 37(1), 91–97. https://doi.org/10.16288/j.yczz.2015.01.013
Chen J, Hu Q, Zhang Y, Lu C, Kuang H (2014) P-MITE: A database for plant miniature inverted-repeat transposable elements. Nucleic Acids Res, 42(Database issue), D1176–1181. https://doi.org/10.1093/nar/gkt1000
Crescente JM, Zavallo D, Helguera M, Vanzetti LS (2018) MITE Tracker: An accurate approach to identify miniature inverted-repeat transposable elements in large genomes. BMC Bioinformatics 19(1):348. https://doi.org/10.1186/s12859-018-2376-y
Dufresne M, Hua-Van A, Abd el Wahab H, M’Barek SB, Vasnier C, Teysset L, Kema GHJ, Daboussi M-J (2007) Transposition of a fungal miniature inverted-repeat transposable element through the action of a Tc1-like transposase. Genetics 175(1):441–452. https://doi.org/10.1534/genetics.106.064360
Feschotte C, Jiang N, Wessler SR (2002) Plant transposable elements: Where genetics meets genomics. Nat Rev Genet 3(5):329–341. https://doi.org/10.1038/nrg793
Grativol C, Thiebaut F, Sangi S, Montessoro P, Santos W da S, Hemerly AS, Ferreira PCG (2019) A miniature inverted-repeat transposable element, AddIn-MITE, located inside a WD40 gene is conserved in Andropogoneae grasses. PeerJ, 7, e6080 https://doi.org/10.7717/peerj.6080
Klai K, Zidi M, Chénais B, Denis F, Caruso A, Casse N, Mezghani Khemakhem M (2022) Miniature inverted-repeat transposable elements (MITEs) in the two Lepidopteran genomes of Helicoverpa armigera and Helicoverpa zea. Insects 13(4):313. https://doi.org/10.3390/insects13040313
Li Y, Zhou G, Ma J, Jiang W, Jin L, Zhang Z, Guo Y, Zhang J, Sui Y, Zheng L, Zhang S, Zuo Q, Shi X, Li Y, Zhang W, Hu Y, Kong G, Hong H, Tan B … Qiu L (2014) De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nature Biotechnol, 32(10), 1045–1052. https://doi.org/10.1038/nbt.2979
Macas J, Neumann P, Požárková D (2003) Zaba: A novel miniature transposable element present in genomes of legume plants. Mol Genet Genomics 269(5):624–631. https://doi.org/10.1007/s00438-003-0869-4
Macko-Podgórni A, Machaj G, Grzebelus D (2021) A global landscape of miniature inverted-repeat transposable elements in the carrot genome. Genes, 12(6), Article 6. https://doi.org/10.3390/genes12060859
Nouroz F, Noreen S, Heslop-Harrison JS (2015) Evolutionary genomics of miniature inverted-repeat transposable elements (MITEs) in Brassica. Mol Genet Genomics 290(6):2297–2312. https://doi.org/10.1007/s00438-015-1076-9
Sampath P, Murukarthick J, Izzah NK, Lee J, Choi H-I, Shirasawa K, Choi B-S, Liu S, Nou I-S, Yang T-J (2014) Genome-wide comparative analysis of 20 miniature inverted-repeat transposable element families in Brassica rapa and B. oleracea. PLOS ONE, 9(4), e94499. https://doi.org/10.1371/journal.pone.0094499
Sherman-Broyles S, Bombarely A, Powell AF et al (2014) The wild side of a major crop: Soybean’s perennial cousins from Down Under. Am J Bot 101:1651–1665. https://doi.org/10.3732/ajb.1400121
Tamura K, Stecher G, Kumar S (2021) MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol 38(7):3022–3027. https://doi.org/10.1093/molbev/msab120
Tang Y, Ma X, Zhao S, Xue W, Zheng X, Sun H, Gu P, Zhu Z, Sun C, Liu F, Tan L (2019) Identification of an active miniature inverted-repeat transposable element mJing in rice. Plant J 98(4):639–653. https://doi.org/10.1111/tpj.14260
Tek AL, Kashihara K, Murata M, Nagaki K (2010) Functional centromeres in soybean include two distinct tandem repeats and a retrotransposon. Chromosome Res 18(3):337–347. https://doi.org/10.1007/s10577-010-9119-x
Venkatesh N, Nandini B (2020) Miniature inverted-repeat transposable elements (MITEs), derived insertional polymorphism as a tool of marker systems for molecular plant breeding. Mol Biol Rep 47(4):3155–3167. https://doi.org/10.1007/s11033-020-05365-y
Wheeler TJ, Eddy SR (2013) nhmmer: DNA homology search with profile HMMs. Bioinformatics (oxford, England) 29(19):2487–2489. https://doi.org/10.1093/bioinformatics/btt403
Xu L, Yuan K, Yuan M, Meng X, Chen M, Wu J, Li J, Qi Y (2020) Regulation of rice tillering by RNA-directed DNA methylation at miniature inverted-repeat transposable elements. Mol Plant 13(6):851–863. https://doi.org/10.1016/j.molp.2020.02.009
Zhuang Y, Wang X, Li X, Hu J, Fan L, Landis JB, Cannon SB, Grimwood J, Schmutz J, Jackson SA, Doyle JJ, Zhang XS, Zhang D, Ma J (2022) Phylogenomics of the genus Glycine sheds light on polyploid evolution and life-strategy transition. Nature Plants, 8(3), Article 3. https://doi.org/10.1038/s41477-022-01102-4
Acknowledgements
This work was partially supported by the TÜBİTAK Project ID 118O670. HYA has a fellowship from the 100/2000 YÖK Program and Ayhan Şahenk Foundation.
Author information
Authors and Affiliations
Contributions
A.L.T. conceived the study and designed the experiments. A.L.T., H.Y.A., and E.C.K. performed the experiments, conducted the data analysis, and wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
13258_2024_1519_MOESM3_ESM.xlsx
Supplementary file3 The physical location and coordinates of GftoMITE1 in the assembled whole genome of Glycine falcata (XLSX 960 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yıldız Akkamış, H., Kaya, E.C. & Tek, A.L. Discovery and genome-wide characterization of a novel miniature inverted repeat transposable element reveal genome-specific distribution in Glycine. Genes Genom (2024). https://doi.org/10.1007/s13258-024-01519-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13258-024-01519-5