Abstract
Cyclodextrin glucanotransferase (CGTase, EC. 2.1.1.19) produced using new alkaliphile Microbacterium terrae KNR 9 has been purified to homogeneity in a single step by the starch adsorption method. The specific activity of the purified CGTase was 45 U/mg compared to crude 0.9 U/mg. This resulted in a 50-fold purification of the enzyme with 33 % yield. The molecular weight of the purified enzyme was found to be 27.72 kDa as determined by SDS-PAGE. Non-denaturing gel electrophoresis and activity staining confirmed the presence of CGTase in crude and the ammonium sulfate precipitate fraction. The purified CGTase has a pI value of 4.2. The optimum pH of 6.0 and 60 °C temperature were found to be the best for CGTase activity. Purified CGTase showed 5.18 kcal/mol activation energy (Ea). The CGTase activity was increased in the presence of metal ions (5 mM): Ca+2 (130 %), Mg+2 (123 %), Mn+2 (119 %) and Co+2 (116 %). The enzyme activity was strongly inhibited in the presence of Hg+2 (0.0 %), Cu+2 (0.0 %) and Fe+2 (3.8 %). Inhibitor N-bromosuccinimide (5 mM) showed the highest 96 % inhibition of CGTase activity. SDS and triton X-100 among different detergents and surfactants (1.0 %, w/v) tested showed 92 % inhibition. Among the organic solvents checked for their effect on enzyme activity, 5 % (v/v) toluene resulted in 48 % increased activity. Polyethylene glycol-6000 showed a 26 % increase in the CGTase activity. The kinetic parameters K m and V max were 10 mg/ml and 146 µmol/mg min, respectively, for purified CGTase.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Amylases are of great significance for biotechnological and industrial applications and have approximately 25 % of the world enzyme market (Reddy et al. 2003). Cyclodextrin glucanotransferase is one of the important members of the α-amylase family 13 of glycosyl hydrolases which can degrade starch. This family of enzymes exhibits diversity in reaction specificities. Amylases generally hydrolyze glycosidic bonds in the starch molecules, but CGTases catalyze transglycosylation as a major reaction with hydrolysis being a minor activity (Van der Veen et al. 2000).
CGTases produce cyclodextrins (CDs) as a result of intramolecular transglycosylation (cyclization) reaction during the degradation of starch. Cyclodextrins are cyclic oligosaccharides commonly composed of six, seven or eight d-glucose units (α, β and γ-cyclodextrins, respectively) joined by α-(1, 4) glycosidic bonds (Szejtli 1998). Because of this unique property, CDs can form molecular inclusion complexes (host–guest complexes) with a wide range of solid, liquid and gaseous compounds and hence affect the stability, solubility, reactivity and bioavailability of a wide range of molecules. So, cyclodextrins have various applications in the field of medicine, food, pharmaceutical, analytical chemistry, agriculture and cosmetics (Hedges 1998). Though CDs have several possible industrial uses, their extensive applications are restricted due to their high prices and low yields (Singh et al. 2002). Hence, screening of novel sources for CGTases has a wide technical and practical impact on the enzymatic production of CDs (Biwer et al. 2002).
The bacillus group of bacteria, e.g., Bacillus coagulans (Akimaru et al. 1991), Bacillus firmus (Goel and Nene 1995), Bacillus agaradhaerens (Martins and Hatti-Kaul 2000), Bacillus lentus (Sabioni and Park 1992), Bacillus ohbensis, Bacillus circulans, Bacillus macerans (Yagi et al. 1986), Bacillus alcalophilus B-3101 (Abelyan et al. 2002) and Bacillus pseudalcaliphilus 20RF (Atanasovva et al. 2011) have been mainly reported for CGTase production. Apart from the Bacillus group, there are reports of CGTase production from other organisms like Klebsiella pneumoniae AS-22 (Gawande and Patkar 2001), Brevibacterium sp. 9605 (Mori et al. 1994), Paenibacillus campinasensis H69-3 (Alves-Prado et al. 2007), Paenibacillus illinoisensis ZY-08 (Lee et al. 2013) Pyrococcus furiosus DSM3638 (Lee et al. 2007), Thermococcus (Tachibana et al. 1999) and Thermoactinomyces vulgaris Tac-5354 (Abelyan et al. 2002). There are also reports of CGTase producing anaerobic bacteria like Thermoanaerobacterium thermosulfurigenes EM1 (Wind et al. 1995) and Anaerobranca gottschalkii (Thiemann et al. 2004). Bautista et al. (2012) reported the CGTase from halophilic archeon Haloferax mediterranei. Though CGTase production and characterization from a variety of aerobic mesophilic bacteria have been reported, there is a need of robust enzymes from other organisms considering the harsh conditions used in starch industries (Wind et al. 1995).
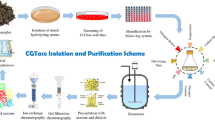
There are many reports on the purification of CGTases using different procedures like ultrafiltration, gel filtration, starch adsorption chromatography, hydrophobic interaction chromatography, ion exchange chromatography and affinity chromatography. We have isolated a new alkaliphile Microbacterium terrae KNR 9 for CGTase production (Rajput et al. 2016). In this study, we report the purification and properties of CGTase produced by Mic. terrae KNR 9.
Materials and methods
Materials
Cornstarch was purchased from Hi-media, Mumbai, India. Ammonium sulfate was bought from Qualigens India Ltd. Standard protein molecular weight markers were procured from Bangalore Genei (GeNeiTM). All other chemicals used were of analytical grade.
Organism and production medium
We have isolated the CGTase producing bacteria from fertile soil (Anand District, Gujarat, India) in our laboratory as described by Park et al. (1989). The highest enzyme-producing bacterial isolate was identified as Microbacterium terrae KNR 9 by IMTECH, Chandigarh, India, and deposited as Microbacterium terrae MTCC 8083 at the same institute. CGTase production was carried out in 100 ml medium containing 20 g/l soluble starch, 10 g/l yeast extract, 1.0 g/l K2HPO4, 0.2 g/l MgSO4·7H2O and 10 g/l Na2CO3 (autoclaved separately) in 250 ml flasks at 30 °C, 150 rpm on a rotary shaker for 72 h. After incubation, cells were removed by centrifugation and the supernatant used for enzyme purification.
CGTase activity and protein estimation
CGTase activity was determined by the phenolphthalein assay method described by Goel and Nene (1995) with minor modification. 100 μl appropriately diluted purified enzyme was incubated with 1.0 ml of 50 mg soluble starch in sodium phosphate buffer (pH 6.0, 50 mM) at 60 °C for 30 min. The reaction was stopped by quickly cooling the tubes on ice. Four milliliters of working phenolphthalein solution was added and the tubes were vortexed. Absorbance of the mixture was immediately measured at 550 nm. The working phenolphthalein solution was prepared by adding 1 ml of phenolphthalein stock (4 mM in ethanol) to 100 ml of 125 mM Na2CO3 containing 4 % ethanol. The amount of β-CD produced was estimated from a standard curve of 0–100 μg/ml pure β-CD obtained by the same procedure. One unit was defined as the amount of enzyme that produced one μmole of β-CD per minute under assay conditions.
Protein estimation was carried out as described by Lowry et al. (1951). The standard protein calibration curve was prepared using pure bovine serum albumin (0–100 μg/ml).
Ammonium sulfate fractionation
Cell-free supernatant was subjected to precipitation by the addition of (ammonium sulfate) (NH4)2SO4 powder to achieve 20 % saturation in an ice bath. Slow and gentle stirring of the mixture was performed for 2 h for better dissolution of ammonium sulfate to promote salting out of proteins. Centrifugation was carried at 9000g for 20 min at 4 °C and a pellet of protein obtained was checked for CGTase activity. Since the pellet did not show any CGTase activity, it was discarded. The supernatant was added with ammonium sulfate to achieve 80 % saturation and kept in an ice bath with gentle stirring for 2 h. The mixture was kept at 4 °C overnight to enhance the precipitation and stabilization of the enzyme. The resulting precipitates were separated from the supernatant by centrifugation at 9000g for 20 min at 4 °C and resuspended in 50 mM phosphate buffer, pH 6.0 and dialyzed against the same buffer at 8 °C for 24 h with three buffer changes. The CGTase activity and protein concentration were measured as mentioned above for the next purification steps.
Purification of CGTase by starch adsorption
We purified the CGTase using the starch adsorption method described by Martins and Hatti-Kaul (2000) with minor modifications. For starch adsorption of CGTase, the crude enzyme was precipitated at 20 % ammonium sulfate saturation and the precipitated protein pellet obtained was discarded and the resultant supernatant of 20 % ammonium sulfate saturation containing soluble proteins subjected to cornstarch adsorption. In the other experiment, crude enzyme was added to plain 5 % (w/v) cornstarch without 20 % (w/v) ammonium sulfate to compare its effect on the adsorption of the enzyme.
To 20 ml crude enzyme, 20 % (w/v) ammonium sulfate was added and kept at 4 °C for 2 h on the stirrer. Then, it was centrifuged at 9000g and 4 °C for 20 min and the supernatant was carefully transferred to another flask discarding the pellet of protein precipitate. To this supernatant with 20 % (w/v) ammonium sulfate, 5 % (w/v) cornstarch was added and kept for 1 h at 8 °C with gentle stirring to allow enzyme adsorption on starch molecules. The mixture was centrifuged at 5000 rpm for 10 min to sediment the cornstarch with adsorbed proteins. The starch sediment obtained was washed twice with 10 ml of cold distilled water to remove unbound proteins. The adsorbed CGTase on the starch was eluted by incubating it with 5 ml of 1 mM β-CD in 50 mM phosphate buffer, pH 6.0, for 30 min at 37 °C on the stirrer. After that it was centrifuged at 9000g and 4 °C for 20 min to remove starch particles. The supernatant containing desorbed CGTase was carefully transferred to another tube. The elution was repeated again with 2.0 ml of the elution buffer. Elutes were pooled and dialyzed against 50 mM phosphate buffer, pH 6.0, at 8 °C for 24 h with three buffer changes. In the other experiment, 20 ml crude enzyme was mixed with 5 % (w/v) cornstarch only and incubated at 8 °C for 1 h with gentle stirring. After that, 5 ml of 1 mM β-CD in 50 mM phosphate buffer, pH 6.0, was added and kept for 30 min at 37 °C on the stirrer to recover adsorbed enzyme.
Characterization of the purified CGTase
Determination of molecular weight
The molecular weight of the purified protein (CGTase) was estimated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). It was performed on a vertical slab gel using 10.0 % (w/v) acrylamide gel at a constant voltage of 125 V. Standard protein molecular weight markers (GeNeiTM) used were phosphorylase B (Mr 97,400 Da); bovine serum albumin (Mr 66,000 Da); ovalbumin (Mr 43,000 Da); carbonic anhydrase (Mr 29,000 Da); soybean trypsin inhibitor (Mr 20,100 Da); and lysozyme (Mr 14,300 Da). The gel was stained by the silver staining method. The molecular weight of the enzyme protein was determined using an Alpha DigiDocTm (Alpha Innotech, California, USA) with the Alpha DigiDoc®RT software.
Non-denaturing gel electrophoresis and activity staining
Native polyacrylamide gel electrophoresis was performed in 8 % gel at a constant voltage of 100 V till the dye front reached the end of the resolving gel. Crude and dialyzed enzyme samples of ammonium sulfate precipitation were analyzed to check the presence of enzyme protein. After completion of the electrophoretic run, the gel was rinsed twice with 50 mM phosphate buffer (pH 6.0).
Activity staining of the gel was done according to Pakzad et al. (2005) with minor modifications. The starch indicator gel was prepared before completion of an electrophoretic run using 0.5 g soluble starch and 0.4 g agar in 20 ml of 50 mM phosphate buffer, pH 6.0, under boiling condition. After that, 0.5 ml of 0.4 g % phenolphthalein was added to this mixture and it was poured in a Petri plate upon cooling to 50 °C. The native gel was carefully transferred by placing it on the starch indicator gel and incubated at 50 °C for 10 min. During incubation, enough care was taken to ensure that the gel remains flooded with a phosphate buffer (50 mM, pH 6.0). After incubation, the native gel was removed carefully and the starch indicator gel flooded with a 0.1 g % sodium carbonate solution until the background of the gel turned pink. The presence of a colorless band in the starch indicator gel confirmed CGTase activity of the separated protein.
After visualization of CGTase activity, the native PAGE gel was subjected to a silver staining method and compared with the starch indicator gel.
Isoelectric focusing
Isoelectric focusing was carried out at 20 °C in the Protean IEF cell (BioRad, USA) according to the manufacturer’s instructions. The isoelectric point of the purified CGTase was analyzed using immobilized pH gradient (IPG) strips (BioRad, USA) with a pH range of 3.0–10.0. Active rehydration of the IPG strip was carried out by keeping it in the solution containing purified CGTase and rehydration buffer at 50 V for 12 h. The rehydration buffer comprised 8 M urea, 2 % CHAPS, 50 mM dithiothreitol (DTT), 0.2 % carrier ampholytes and 0.001 % Bromophenol Blue. After rehydration of the IPG strip, focusing was carried out on Protean IEF cell (BioRad, USA) initially at 250 V for 15 min, 10,000 V for the next 3 h and the final focusing was done till 40,000 Vh was achieved. The enzyme protein was visualized by silver staining of the IPG strip.
Effect of pH on CGTase activity
The effect of pH on CGTase activity was checked using the phenolphthalein assay method as described earlier. The optimum pH of the purified CGTase enzyme was determined by replacing 50 mM phosphate buffer (pH 6.0) with the followings buffers: 50 mM sodium acetate buffer (pH 4.0, 5.0 and 5.5), 50 mM sodium phosphate buffer (pH 6.0, 6.5 and 7.0), 50 mM Tris–HCl (pH 8.0) and 50 mM glycine–NaOH buffer (pH 9.0, 10.0 and 10.5). Considering the enzyme activity at the best pH as 100 %, a pH profile of the relative activity versus pH was plotted.
Effect of temperature on CGTase activity
The effect of temperature on enzyme activity was tested at different temperatures. The optimum temperature for CGTase activity was determined by incubating the reaction mixture of CGTase assay at different temperatures, ranging from 30 to 70 °C in 50 mM phosphate buffer (pH 6.0) for 20 min. Considering the enzyme activity at the best temperature as 100 %, a temperature profile of the relative activity versus temperature was plotted.
Effect of metal ions on CGTase activity
Effect of different metal salts, namely CaCl2, MgSO4, FeSO4, ZnSO4, CuSO4, MgCl2, MnCl2, ZnCl2, CoCl2, HgCl2, NiCl2, K2Cr2O7 and AgNO3 on CGTase enzyme activity was determined. Appropriately diluted 100 µl of enzyme was mixed with metals (5 mM final concentration) and incubated at 25 °C for 10 min. The residual activity of the enzyme was determined.
Effect of inhibitors on CGTase activity
The effect of different inhibitors, namely p-chloromercuribenzoic acid (p-CMB), phenylmethylsulfonyl fluoride (PMSF), N-bromosuccinimide (NBS), dithiothreitol (DTT), ethylenediamine tetraacetate (EDTA) and β-mercaptoethanol (β-ME), on CGTase activity was checked. Appropriately diluted 100 µl of an enzyme was mixed with the respective inhibitors (5.0 mM final concentration) and incubated at 25 °C for 10 min. Residual enzyme activity was determined.
Effect of detergents on CGTase activity
Effect of various detergents (1.0 %, w/v), namely, cetyl trimethyl ammonium bromide (C-TAB), sodium dodecyl sulfate (SDS), Tween 40, Tween 80 and Triton X-100 on CGTase activity was checked. The approximately diluted 100 µl enzyme was mixed with the detergents and incubated at 25 °C for 10 min. After that, the residual enzyme activity was determined by the CGTase assay.
Effect of organic solvents on CGTase activity
Effect of various organic solvents, viz. acetone, ethanol, isopropanol, hexane, cyclohexane, toluene, iso-octane and dodecane, on CGTase activity was checked. Organic solvents (5 %, v/v) were added to the tubes containing substrate (0.5 g % soluble starch) and approximately diluted 100 µl enzyme. The reaction mixture was incubated at 25 °C for 10 min followed by the CGTase assay method.
Effect of polyols on CGTase activity
The effect of different polyols, viz. glycerol, mannitol, sorbitol and polyethylene glycol, on CGTase activity was determined. Different polyols (0.5 M final concentration) were mixed with soluble starch (0.5 g % final concentration) and then appropriately diluted 100 µl enzyme was added to it. The reaction mixtures were incubated at 60 °C for 20 min followed by the CGTase assay method.
Kinetic parameters
K m and V max values for the purified CGTase enzyme were determined. Fixed 0.27 µg of purified CGTase protein was incubated with different soluble starch concentrations ranging from 0.5 to 5.0 mg/ml in 1.0 ml of 50 mM phosphate buffer, pH 6.0 at 60 °C for 4 min. The values of K m and V max were determined from a Lineweaver–Burk plot.
Results and discussion
Concentration by ammonium sulfate precipitation
CGTase production was performed as mentioned earlier and the cell-free supernatant, i.e., crude enzyme was used for further purification. This crude CGTase enzyme was first concentrated by precipitation following ammonium sulfate fractionation. The protein pellet of 0–20 % showed no CGTase activity. The 20–80 % salt saturation concentrated protein pellet showed 9.84 U/mg of specific CGTase activity with 72 % yield (Table 1).
Purification of CGTase by starch adsorption
The CGTase from Mic. terrae KNR 9 was purified to homogeneity by adsorption onto cornstarch followed by elution with buffer containing β-cyclodextrin (Martins and Hatti-Kaul 2000). The precipitation and adsorption of CGTase were promoted in the presence of 20 % ammonium sulfate. In an experiment where plain cornstarch was used directly for the adsorption of the crude enzyme, only 9.31 % yield was obtained. In the other method, crude enzyme was precipitated at 20 % ammonium sulfate saturation and the precipitated protein pellet obtained was discarded. The resultant supernatant of 20 % ammonium sulfate saturation containing soluble proteins was subjected to cornstarch adsorption. By this method, CGTase from Mic. terrae KNR 9 could be purified to homogeneity in a single step with 45.22 U/mg specific activity, 50-fold purification and 33 % yield (Table 1).
Martins and Hatti-Kaul (2000) reported purification of CGTase from B. agaradhaerens LC-3C using the starch adsorption method with 50 % yield and 43-fold purification. Rosso et al. (2005) described the CGTase purification from B. circulans using the starch adsorption method. Vassileva et al. (2007) obtained purified CGTase with 87 % enzyme yield and 1.3-fold purification.
Characterization of CGTase from Mic. terrae KNR 9
Molecular weight determination
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) is a widely used method for identification, screening and assessment of homogeneity of the purified protein fractions. The purified CGTase exhibited a single protein band on the 10 % polyacrylamide gel stained by silver indicating that it was a monomer protein (Fig. 1). The molecular weight of the purified enzyme was assessed by electrophoretic mobility of proteins in the denaturing SDS-PAGE gel. The molecular weight of CGTase from Mic. terrae KNR 9 was 27.72 kDa compared to the distance traveled by individual standard molecular weight markers using the Alpha DigiDoc®RT software. To the best of our knowledge, this is the smallest molecular weight CGTase reported so far.
The other CGTases reported having lower molecular weights are B. coagulans, 36 kDa (Akimaru et al. 1991), and B. lentus, 33 kDa (Sabioni and Park 1992). The majority of purified CGTases reported in the literature have a molecular weight between 65 and 80 kDa, e.g., Bacillus pseudalcaliphilus 20RF, 70 kDa (Atanasovva et al. 2011), Paenibacillus campinasensis strain H69-3, 70 kDa (Alves-Prado et al. 2007), Paenibacillus illinoisensis ZY-08, 74 kDa (Lee et al. 2013), B. stearothermophilus ET1, 66.88 kDa, (Chung et al. 1998), Bacillus firmus, 78 kDa (Gawande et al. 1999), Bacillus pseudalcaliphilus 8SB, 71 kDa, (Kitayska et al. 2011) and Bacillus megaterium 73.4 kDa (Pishtiyski et al. 2008). On the other hand, CGTase from B. agaradhaerens LC-3C has a higher molecular weight of 110 kDa (Martins and Hatti-Kaul 2000). Abelyan et al. (2002) noticed that CGTases of alkaliphilic strains had higher molecular weight. In contrast to this, CGTase from new alkaliphile Mic. terrae KNR 9 has a lower molecular weight of 27.72 kDa.
Non-denaturing gel electrophoresis and activity staining
The presence of CGTase was confirmed by non-denaturing gel electrophoresis followed by activity staining as described earlier. Two colorless bands appeared upon addition of 0.1 % (w/v) sodium carbonate in the pink-colored starch indicator gel indicating CGTase activity (Fig. 2). The corresponding two bands were observed in the non-denaturing gel after silver staining in crude and ammonium sulfate-concentrated enzyme lane. CGTase is an enzyme of the α-amylase family and shows starch degradation as like other amylases and can be detected by the addition of iodine solution on starch gel after native PAGE (Alves-Prado et al. 2007; Thiemann et al. 2004). However, it indicates starch hydrolysis only and cannot discriminate between two amylolytic enzymes. Activity staining with this specific phenolphthalein starch indicator gel method reveals only bands of CGTase activity (Pakzad et al. 2005).
Isoelectric focusing
The CGTase from Mic. terrae KNR 9 showed isoelectric pH (pI) of 4.2. Mori et al. (1994) and Yagi et al. (1986) also reported the acidic pI of 2.8 and <4.0 for CGTase from Brevibacterium sp. 9605 and B. ohbensis, respectively. The other reported pIs for CGTases are Pae. campinasensis H69-3, 5.3 (Alves-Prado et al. 2007), B. agaradhaerens LC-3C, 6.9 (Martins and Hatti-Kaul 2002), K. pneumoniae AS-22, 7.3 (Gawande and Patkar 2001), B. circulans IFO 3329, 8.8 (Yagi et al. 1986) and Bacillus sp. G1, 8.8 (Sian et al. 2005).
Effect of pH on CGTase activity
Enzyme activity was measured in different buffers between pH 4.0 to 10.5 at 60 °C using the CGTase assay method. The best enzyme activity was observed at 6.0 pH (50 mM phosphate buffer) and so it is the optimum pH for the purified enzyme (Fig. 3). The optimal pH value obtained is in accordance with other reported pH values in the literature. The CGTases from B. circulans ATCC 21783 (Vassileva et al. 2007), B. stearothermophilus ET1 (Chung et al. 1998), Bacillus sp. G1 (Sian et al. 2005), B. firmus No. 37 (Matioli et al. 2001) and B. firmus (Moriwaki et al. 2007) have shown the highest enzyme activity at 6.0 pH.
The CGTase from Mic. terrae KNR 9 showed no activity at pH ≤4.0 and ≥10.5. Extreme pH values were not found suitable for CGTase activity. It needs near-neutral pH for its highest activity. However, CGTase from Mic. terrae KNR 9 showed another broad peak of 60 % relative activity at 8.0–9.0 pH (Fig. 3). The other reported CGTases having near-neutral optimum pH are B. coagulans (Akimaru et al. 1991), K. pneumoniae AS-22 (Gawande and Patkar 2001), Pae. campinasensis H69-3 (Alves-Prado et al. 2007), Pae. illinoisensis ZY-08 (Lee et al. 2013), Thermococcus sp. (Tachibana et al. 1999), B. pseudalcaliphilus 8SB (Kitayska et al. 2011), B. megaterium (Pishtiyski et al. 2008), B. firmus (Yim et al. 1997) and B. lehensis (Elbaz et al. 2015).
Effect of temperature on CGTase activity
The CGTase activity was determined at different temperatures ranging from 30 to 70 °C. The temperature of 60 °C showed the highest activity for the purified CGTase from Mic. terrae KNR 9 (Fig. 4). Similarly, a temperature of 60 °C was found to be optimum for CGTases of B. megaterium (Pishtiyski et al. 2008), B. circulans E192 (Bovetto et al. 1992) and B. lehensis (Elbaz et al. 2015). Most of the CGTases reported have optimum temperature between 50 and 65 °C for their activity (Atanasovva et al. 2011; Lee et al. 2007; Martins and Hatti-Kaul 2002; Matioli et al. 2001; Vassileva et al. 2007; Yim et al. 1997).
The higher optimum temperature of 75 °C has been noted for CGTases from B. alcalophilus B-3101 and Thermoactinomyces vulgaris Tac-5354 (Alves-Prado et al. 2007). There are reports of thermostable CGTases from B. stearothermophilus at 80 °C (Chung et al. 1998) and Thermoanaerobacter thermosulfurigenes EM1 at 80–85 °C (Wind et al. 1995). In contrast to this, Lee et al. (2013) noticed 40 °C as the best for CGTase activity of Pae. illinoisensis ZY-08.
Activation energy of CGTase from Mic. terrae KNR 9
The effect of temperature on an enzymatic reaction can be analyzed through the Arrhenius equation by plotting the velocity (log10V) against the inverse of temperature (1/T expressed in K−1) (Fig. 5). For purified CGTase, the activation energy (Ea) determined was 5.18 kcal/mol. Reported Ea for other different CGTases are B. agaradhaerens LC-3C, 17 kcal/mol (Martins and Hatti-Kaul 2002), B. firmus no. 37, 7.5 kcal/mol (Matioli et al. 2001) and B. firmus, 9.4 kcal/mol (Moriwaki et al. 2007).
Effect of metal ions on CGTase activity
To check the effect of metal ions on purified CGTase, the enzyme was incubated with some metal salts. Purified CGTase was added to metal salts 5.0 mM, in 50 mM phosphate buffer, pH 6.0 at 25 °C for 10 min and then the residual activity was determined. The results of CGTase activity are summarized in Table 2. The metal ions Hg+2 and Cu+2 completely inhibited the CGTase of Mic. terrae KNR 9, showing no residual activity (0.0 %). The purified CGTase was also strongly suppressed by Fe+2 and Ag+3 giving 3.84 % and 14.10 % residual activity, respectively. The CGTase was slightly inhibited in the presence of Zn+2 and Cr+3 showing about 90 % residual activity. No effect of Ni+2 was observed on CGTase activity. The CGTase activity was slightly enhanced in the presence of Ca+2, Mg+2, Co+2 and Mn+2.
A strong inhibition of CGTase activity with Hg+2 has been reported by Akimaru et al. (1991) and Martins and Hatti-Kaul (2002). There are reports of significant inhibitory effect of Cu+2 on CGTases of various bacteria (Akimaru et al. 1991; Mori et al. 1994; Kitayska et al. 2011; Martins and Hatti-Kaul 2002; Sian et al. 2005; Yim et al. 1997). Similar inhibitory effects of Fe+2 were reported for CGTase from Bacillus sp. AL-6 (Fujita et al. 1990) and B. agaradhaerens LC-3C (Martins and Hatti-Kaul 2002). Similar to CGTase from Mic. terrae KNR 9, CGTase from B. megaterium (Pishtiyski et al. 2008), Bacillus sp. G1 (Sian et al. 2005) and Bacillus sp. AL-6 (Fujita et al. 1990) showed inhibition with Zn+2. Most of the reported CGTases showed either enhanced CGTase activity and stability or retained the activity in the presence of Ca+2 (Akimaru et al. 1991; Mori et al. 1994; Lee et al. 2013; Kitayska et al. 2011; Martins and Hatti-Kaul 2002; Sian et al. 2005; Yim et al. 1997). The improvement in the activity could be related to the interactions of Ca+2 ions with the amino acids near the active site that are conserved among the α-amylase family enzymes (Martins and Hatti-Kaul 2002).
Effect of inhibitors on CGTase activity
The effect of different known enzyme inhibitors, viz. ethylenediamine tetraacetate (EDTA), phenylmethylsulfonylfluoride (PMSF), dithioerythritol (DTT), β-mercapto-ethanol (β-ME), N- bromosuccinimide (NBS) and p-chloromercuribenzoate (p-CMB), on the CGTase activity was checked (Table 3).
N-Bromosuccinimide showed 96 % inhibition of the CGTase activity. It is known to oxidize the tryptophan residues present in the enzyme and thereby inhibit the enzyme activity (Spende et al. 1966). Hence, it can be stated that the tryptophan and related amino acids might be present in the active site of purified CGTase and have a crucial role in enzyme action. Similar results were reported with CGTase from B. agaradhaerens LC-3C (Martins and Hatti-Kaul 2002). Close to 40 % inhibition of enzyme activity was observed with p-CMB, indicating the presence of the sulfhydryl groups on the active site of the enzyme. None of the other inhibitors tested, i.e., EDTA, PMSF, DTT and β-ME, showed significant inhibition of CGTase activity. Similar results for the effect of inhibitors for CGTases from other sources have been reported (Akimaru et al. 1991; Fujita et al. 1990; Martins and Hatti-Kaul 2002; Yim et al. 1997).
Effect of detergents on CGTase activity
The effect of several detergents/surfactants on CGTase activity is summarized in Table 4. The highest inhibition of 92 % CGTase activity was observed with a nonionic detergent Triton X-100. Anionic surfactants SDS and CTAB also showed higher inhibition of 91 and 75 %, respectively. Nonionic surfactants Tween-40 and Tween-80 revealed about 30 % inhibition of enzyme activity.
Effect of organic solvents on CGTase activity
The effect of various organic solvents, viz. acetone, ethanol, isopropanol, hexane, cyclohexane, toluene, iso-octane and dodecane, on cyclodextrin production by CGTase is represented in Table 5. Among the tested organic solvents, toluene, dodecane, hexane, cyclohexane and iso-octane increased the cyclodextrin yield, whereas acetone and isopropanol showed decreased cyclodextrin production. The addition of toluene to the reaction mixture resulted in the highest 48 % increase of CGTase activity, followed by dodecane, hexane, cyclohexane and iso-octane.
Abelyan et al. (2002) observed increased β-CD yield using various organic solvents with toluene showing the highest impact. Blackwood and Bucke (2000) reported the increase in cyclodextrin yield with CGTases from Thermoanaerobacter sp. and B. circulans 251 using different solvents. Qi et al. (2007) noticed the effect of ethanol on the yield of large-ring CDs using CGTase from Bacillus sp. BT3-2. Polar organic solvents can form inclusion complexes with cyclodextrins and may result in conformational changes in the cyclodextrin structure which can decrease product inhibition.
Effect of polyols on CGTase activity
The effect of different polyols at 0.5 M final concentration was determined on CGTase activity and the results are summarized in Table 6. Among the polyols tested, only PEG-6000 showed positive effect on CGTase with 26 % increased activity. Rest of the tested polyols have no effect or slightly inhibitory effect on CGTase activity.
Martins and Hatti-Kaul (2002) noticed the increased activity with sorbitol and PEG-3000. Delbourg et al. (1993) reported the improved activity in the presence of glycerol, mannitol and PEG-200, but decreased activity with xylitol and sorbitol. Addition of polyethylene glycol and polypropylene glycol increased the CD production by CGTase of B. ohbensis (Hayashida and Kawakami 1992). Yoon and Robyt (2005) have reported 20 % increased activity with PEG-1500 for CGTase from B. macerans. Cyclodextrin production may be influenced by factors causing changes in the substrate/enzyme or additive/active site interactions, or even with changes in the microenvironment of the enzyme (e.g., reduction of water activity) induced by the additive (Martins and Hatti-Kaul 2002).
Kinetic parameters
The kinetic parameters for the CGTase catalyzed reaction were determined under standard assay conditions using starch as substrate. K m and V max values obtained for purified CGTase were 10 mg/ml and 146 µmol/mg min, respectively (Fig. 6).
The higher value of K m indicates the lower affinity for the substrate and hence the reaction rate. The CGTase from Pae. campinasensis H69-3 has the Km and V max value of 1.69 mg/ml and 4.97 µmol/min mg, respectively (Alves-Prado et al. 2007). The K m and V max values were 0.48 mg/ml and 51.38 mg/ml min, respectively, for Paenibacillus illinoisensis ZY-08 CGTase (Lee et al. 2013). The values of K m 14.3 g/l and V max 106 U/mg were reported using potato starch as substrate for Haloferax mediterranei (Bautista et al. 2012). The K m of 21.2 mg/ml and V max of 7.4 µmol/mg min have been reported for the CGTase from B. agaradhaerens LC-3C (Martins and Hatti-Kaul 2002). The K m and V max values obtained were 0.15 mg/ml and 60.39 mg/ml min, respectively, for Bacillus sp. G1 CGTase (Sian et al. 2005). K m 2.2 mg/ml and V max 7.8 mg/ml min were reported for CGTase of B. lehensis (Elbaz et al. 2015).
Conclusion
The present study describes the single step purification of cyclodextrin glucanotransferase using the starch adsorption method. The purified CGTase with 45.22 U/mg specific activity gave a single band in SDS-PAGE. To the best of our knowledge, this is the smallest molecular weight CGTase with 27.72 kDa reported so far. The purified enzyme works efficiently near neutral pH and at 60 °C temperature. The CGTase activity can be enhanced in the presence of Ca+2 and other metal ions. It remains active in several organic solvents with toluene as the best. The addition of PEG-6000 can increase the enzyme activity. These diverse properties of novel CGTase can be potentially exploited for industrial cyclodextrin production.
References
Abelyan VA, Balayan AM, Manukyan LS, Afyan KB, Melicsetyan VS, Andreasyan NA, Markosyan AA (2002) Characteristics of cyclodextrin production using cyclodextrin glucanotransferase from various groups of microorganisms. Appl Biochem Microbiol 38:527–535
Akimaru K, Yagi T, Yamamoto S (1991) Purification and properties of Bacillus coagulans cyclomaltodextrin glucanotransferase. J Ferment Bioeng 71:322–328
Alves-Prado HF, Gomes E, Da Silva R (2007) Purification and characterization of a cyclomaltodextrin glucanotransferase from Paenibacillus campinasensis strain H69-3. Appl Biochem Biotechnol 136–140:41–56
Atanasovaa N, Kitayskaa T, Bojadjievaa I, Yankovb D, Tonkova A (2011) A novel cyclodextrin glucanotransferase from alkaliphilic Bacillus pseudalcaliphilus 20RF: purification and properties. Process Biochem 46:116–122
Bautista V, Esclapez J, Pérez-Pomares F, Martínez-Espinosa RM, Camacho M, Bonete MJ (2012) Cyclodextrin glycosyltransferase: a key enzyme in the assimilation of starch by the halophilic Archaeon Haloferax mediterranei. Extremophiles 16:147–159
Biwer A, Antranikian G, Heinzle E (2002) Enzymatic production of cyclodextrins. Appl Microbiol Biotechnol 59:609–617
Blackwood A, Bucke C (2000) Addition of polar organic solvents can improve the product selectivity of cyclodextrin glycosyltransferase—solvent effects on CGTase. Enz Microbiol Technol 27:704–708
Bovetto LJ, Backer DP, Villette JR, Sicard PJ, Bouquelet SJ (1992) Cyclomaltodextrin glucanotransferase from Bacillus circulans E 192: purification and characterization of the enzyme. Biotechnol Appl Biochem 15:48–5838
Chung HJ, Yoon SH, Lee MJ, Kim MJ, Kweon KS, Lee IW, Kim JW, Oh BH, Lee HS, Spiridonova VA, Park KW (1998) Characterization of a thermostable cyclodextrin glucanotransferase isolated from Bacillus stearothermophilus ET1. J Agricult Food Chem 46:952–959
Delbourg MF, Drouet PH, Moraes FD, Thomas D, Barbotin JN (1993) Effect of PEG and other additives on cyclodextrin production by Bacillus macerans cyclomaltodextrin-glycosyl-transferase. Biotechnol Lett 15:157–162
Elbaz A, Sobhi A, ElMekawy A (2015) Purification and characterization of cyclodextrin β-glucanotransferase from novel alkalophilic bacilli. Bioprocess Biosyst Eng 38:767–776
Fujita Y, Tsubouchi H, Inagi Y, Tomita K, Ozaki A, Nakanishi K (1990) Purification and properties of cyclodextrin glycosyltransferase from Bacillus sp. AL-6. J Ferment Bioeng 70:150–154
Gawande BN, Patkar AY (2001) Purification and properties of a novel raw starch degrading-cyclodextrin glycosyltransferase from Klebsiella pneumoniae AS-22. Enz Microbiol Technol 28:735–743
Gawande BN, Goel A, Patkar AY, Nene SN (1999) Purification and properties of a novel raw starch degrading cyclomaltodextrin glucanotransferase from Bacillus firmus. Appl Microbiol Biotechnol 51:504–509
Goel A, Nene S (1995) A novel cyclomaltodextrin glucanotransferase from Bacillus firmus that degrades raw starch. Biotech Lett 17:411–416
Hayashida H, Kawakami (1992) Enhancement of enzymatic production of cyclodextrins by adding polyethylene glycol or polypropylene glycol. J Ferment Bioeng 73(3):239–240
Hedges AR (1998) Industrial applications of cyclodextrins. In Chem Rev 98:2035–2044
Kitayska T, Petrova P, Ivanova V, Tonkova A (2011) Purification and properties of a new thermostable cyclodextrin glucanotransferase from Bacillus pseudalcaliphilus 8SB. Appl Biochem Biotechnol 165:1285–1295
Lee MH, Yang SJ, Kim JW, Lee HS, Kim JW, Park KH (2007) Characterization of a thermostable cyclodextrin glucanotransferase from Pyrococcus furiosus DSM3638. Extremophiles 11:537–541
Lee YS, Zhou Y, Park DJ, Chang J, Choi YL (2013) β-cyclodextrin production by the cyclodextrin glucanotransferase from Paenibacillus illinoisensis ZY-08: cloning, purification, and properties. World J Microbiol Biotechnol 29:865–873
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275
Martins RF, Hatti-Kaul R (2000) A new cyclodextrins glycosyltransferase from an alkaliphilic Bacillus agaradhaerens isolate: purification and characterisation. Enz Microbiol Technol 30:116–124
Martins RF, Hatti-Kaul R (2002) Bacillus agaradhaerens LS-3C cyclodextrin glycosyltransferase: activity and stability features. Enz Microbiol Technol 33:819–827
Matioli G, Zanin GM, De Moraes FF (2001) Characterization of cyclodextrin glycosyltransferase from Bacillus firmus strain no. 37. Appl Biochem Biotechnol 91–93:643–654
Mori S, Hirose S, Oya T, Kitahata S (1994) Purification and properties of cyclodextrin glucanotransferase from Brevibacterium sp. No. 9605. Biosci Biotechnol Biochem 58:1968–1972
Moriwaki C, Costa GL, Pazetto R, Zanin GM, Moraes FF, Portilho M, Matioli G (2007) Production and characterization of a new cyclodextrin glycosyltransferase from Bacillus firmus isolated from Brazilian soil. Process Biochem 42:1384–1390
Pakzad SR, Ajdary S, Moazami N, Haghighi S (2005) A novel method to detect β-cyclodextrin glucosyltransferase (β-CGTase) activity on polyacrylamide gels. Iran Biomed J 9:87–90
Park CS, Park KH, Kim SH (1989) A rapid screening method for alkaline β-cyclodextrin glucanotransferase using phenolphthalein-methyl orange containing solid medium. Agric Biolog Chem 53:1167–1169
Pishtiyski I, Popova V, Zhekova B (2008) Characterization of cyclodextrin glucanotransferase produced by Bacillus megaterium. Appl Biochem Biotechnol 144:263–272
Qi Q, Mokhtar MN, Zimmermann W (2007) Effect of ethanol on the synthesis of large-ring cyclodextrins by cyclodextrin glucanotransferases. J Incl Phenom Macrocycl Chem 57(1):95–99
Rajput KN, Patel KC, Trivedi UB (2016) Screening and selection of medium components for cyclodextrin glucanotransferase production by new alkaliphile Microbacterium terrae KNR 9 using Plackett–Burman Design. Biotechnol Res Int. doi:10.1155/2016/3584807 (Article ID 3584807)
Reddy NS, Nimmagadda A, Rao KRS (2003) An overview of the microbial α-amylase family. Afri J Biotechnol 2–12:645–648
Rosso A, Ferrarotti S, Miranda MV, Krymkiewicz N, Nudel BC, Cascone O (2005) Rapid affinity purification processes for cyclodextrin glycosyltransferase from Bacillus circulans. Biotech Lett 27:1171–1175
Sabioni JG, Park YK (1992) Cyclodextrin glycosyltransferase production by alkaliphilic Bacillus lentus. Rev Microbiol 23:128–132
Sian KH, Said M, Hassan O, Kamarruddin K, Ismail AF, Rahman RA, Mahmood NAN, Illias RM (2005) Purification and characterization of cyclodextrin glucanotransferase from alkaliphilic Bacillus sp. G1. Process Biochem 40:1101–1111
Singh M, Sharma R, Banerjee U (2002) Biotechnological applications of cyclodextrins. Biotechnol Adv 20:341–359
Spande TF, Green NM, Witkop B (1966) The reactivity towards N- bromosuccinimide of tryptophan in enzymes, zymogens and inhibited enzymes. Biochemistry 5:1926–1933
Szejtli J (1998) Introduction and general overview of cyclodextrin chemistry. Chem Rev 98:1743–1753
Tachibana Y, Kuramura A, Shirasaka N, Suzuki Y, Yamamoto T, Fujiwara S, Takagi M, Imanaka T (1999) Purification and characterization of an extremely thermostable cyclomaltodextrin glucanotransferase from a newly isolated hyperthermophilic archeon a Thermococcus sp. Appl Environ Microbiol 65:1991–1997
Thiemann V, Donges C, Prowe SG, Sterner R, Antranikian G (2004) Characterisation of a thermoalkali-stable cyclodextrin glycosyltransferase from the anaerobic thermoalkaliphilic bacterium Anaerobranca gottschalkii. Arch Microbiol 182:226–235
Van der Veen BA, Alebeek GWM, Uitdehaag JCM, Dijkstra BW, Dijkhuizen L (2000) The three transglycosylation reactions catalyzed by cyclodextrin glycosyltransferase from Bacillus circulans (strain 251) proceed via different kinetic mechanisms. Euro J Biochem 267:658–665
Vassileva A, Atanasova N, Ivanova V, Dhulster P, Tonkova A (2007) Characterisation of cyclodextrin glucanotransferase from Bacillus circulans ATCC 21783 in terms of cyclodextrin production. Ann Microbiol 57:609–615
Wind RD, Liebl W, Buitelaar RM, Penninga D, Spreinat A, Dijkhuizen L, Bahl H (1995) Cyclodextrin formation by the thermostable alpha-amylase of Thermoanaerobacterium Thermosulfurigenes EM1 and reclassification of the enzyme as a cyclodextrin glycosyltransferase. Appl Environ Microbiol 61:1257–1265
Yagi Y, Sato M, Ishikura T (1986) Comparative studies of CGTase from Bacillus ohbensis, Bacillus macerans and Bacillus circulans, and production of cyclodextrins using those CGTases. J Jap Soc Starch Sci 33:144–151
Yim DG, Sato HH, Park YH, Park YK (1997) Production of cyclodextrin from starch by cyclodextrin glycosyltransferase from Bacillus firmus and characterization of purified enzyme. J Ind Microbiol Biotechnol 18:402–405
Yoon SH, Robyt JF (2005) Activation and stabilization of 10 starch degrading enzymes by triton X-100, polyethylene glycols, and polyvinyl alcohols. Enz Microbiol Technol 37:556–562
Acknowledgments
Kiransinh Rajput is thankful to the University Grants Commission, New Delhi, for providing teachers fellowship during the course of the study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Rajput, K.N., Patel, K.C. & Trivedi, U.B. A novel cyclodextrin glucanotransferase from an alkaliphile Microbacterium terrae KNR 9: purification and properties. 3 Biotech 6, 168 (2016). https://doi.org/10.1007/s13205-016-0495-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-016-0495-6