Abstract
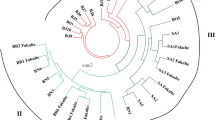
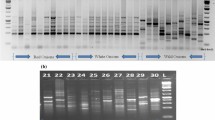
Euryale ferox Salisbury is an important aquatic food plant cultivated largely in eastern India. E. ferox is a monotypic genus, and breeding programmes have mostly relied on the variability present in the primary gene pool. Knowledge of the genetic structure of the population is limited, and there are very few reports available on the genetic diversity of E. ferox. In this study, comprehensive research on the genetic diversity of 16 germplasms of E. ferox was carried out using random amplified polymorphic DNA (RAPD) and inter-simple sequence repeat (ISSR) markers. Out of 320 RAPD and 95 ISSR primers screened initially, 61 primers (40 RAPD and 21 ISSR) gave reproducible bands and were selected for further work. Amplification of the 40 RAPD primers gave 533 polymorphic bands with an average of 13.32 polymorphic bands per primer. The percentage of polymorphism ranged from 37.5 to 100, with an average of 88.3 %. The 21 ISSR primers produced 259 bands, of which 214 were polymorphic, with an average of 10.19 polymorphic bands per primer. The percentage of polymorphism using ISSR primers ranged from 50 to 100, with a mean of 82.6 %. Jaccard’s coefficient ranged from 0.45 to 0.69 (RAPD), 0.50 to 0.77 (ISSR) and 0.48 to 0.71 (RAPD and ISSR). Molecular characterization of different germplasms of E. ferox not only is essential for its conservation but also can be used in further breeding programmes.



Similar content being viewed by others
References
Mandal, R. N., Saha, G. S., & Sarangi, N. (2010). Harvest and processing of Makhana (Euryale ferox Salisb.)—an unique assemblage of traditional knowledge. Indian Journal of Traditional Knowledge, 9, 684–688.
Imanishi, A., Kaneko, S., Isagi, Y., Imanishi, J., Natuhara, Y., & Morimoto, Y. (2011). Development of microsatellite markers for Euryale ferox (Nymphaeaceae), an endangered aquatic plant species in Japan. American Journal of Botany, 98, 233–235.
Jha, V., & Barak, G. K. (2003). Nutritional and medicinal properties of Euryale ferox Salisbury. In M. RK, V. Jha, & D. PV (Eds.), Makhana (pp. 230–238). New Delhi: ICAR.
Nath, B. K., & Chakraborty, A. K. (1985). Studies on amino acid composition of the seeds of Euryale ferox Salisb. Journal of Food Science and Technology, 22, 293.
Das, S., Der, P., Raychaudhuri, U., Maulik, N., & Das, D. (2006). The effect of Euryale ferox (makhana), an herb of aquatic origin, on myocardial ischemic reperfusion injury. Molecular and Cellular Biochemistry, 289, 55–63.
Zhao, H., & Zhao, S. (1994). New cerebrosides from Euryale ferox. Journal of Natural Products, 57, 138–141.
Soller, M., & Beckmann, J. S. (1983). Genetic polymorphism in varietal identification and genetic improvement. Theoretical and Applied Genetics, 67, 25–33.
Welsh, J., & McClelland, M. (1990). Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Research, 18, 7213–7218.
Pamidiamarri, D. V. N. S., Pandya, N., Reddy, M. P., & Radhakrishnan, T. (2009). Comparative study of interspecific genetic divergence and phylogenic analysis of genus Jatropha by RAPD and AFLP. Molecular Biology Reports, 36, 901–907.
Rathore, M. S., Chikara, J., Mastan, S. G., Rahman, H., & Vijyanad, K. G. (2011). Assessment of genetic stability and instability of tissue culture-propagated plantlets of Aloe vera L. by RAPD and ISSR markers. Applied Biochemistry and Biotechnology, 165, 1356–1365.
Gajera, B. B., Kumar, N., Singh, A. S., Punvar, B. S., et al. (2010). Assessment of genetic diversity in castor (Ricinus communis L.) using RAPD and ISSR markers. Industrial Crops and Products, 32, 491–498.
Desai, P., Gajera, B., Mankad, M., Shah, S., Patel, A., Patil, G., Naratan, S., & Kumar, N. (2015). Comparative assessment of genetic diversity among Indian bamboo genotypes using RAPD and ISSR markers. Molecular Biology Reports, 42, 1265–1273.
Rathore, M. S., Yadav, P., Mastan, S. G., Prakash, C. R., Singh, A., & Agarwal, P. K. (2014). Evaluation of genetic homogeneity in tissue culture regenerates of Jatropha curcas L. using flow cytometer and DNA-based molecular markers. Applied Biochemistry and Biotechnology, 172, 1–13.
Rathore, M. S., Mastan, S. G., Yadav, P., Bhatt, V. D., Shekhawat, N. S., & Chikara, J. (2016). Shoot regeneration from leaf explants of Withania coagulans (Stocks) Dunal and genetic stability evaluation of regenerates with RAPD and ISSR markers. South African Journal of Botany, 102, 12–17.
Lin, X. C., Ruan, X. S., Lou, Y. F., Guo, X. Q., & Fang, W. (2008). Genetic similarity among cultivars of Phyllostachys pubescens. Plant Systematics and Evolution, 277, 67–73.
Li, A., & Ge, S. (2001). Genetic variation and clonal diversity of Psammochloa villosa (Poaceae) detected by ISSR markers. Annals of Botany, 87, 585–590.
Quan, Z., Pan, I., Ke, W., & Ding, Y. (2009). Polymorphic microsatellite markers in Euryale ferox Salisb. (Nymphaeaceae). Molecular Ecology Resources, 9, 330–332.
Doyle, J. J., & Doyle, J. L. (1990). Isolation of plant DNA from fresh tissue. Focus, 12, 13–15.
Williams, J. G. K., Kubelik, A. R., Livak, K. J., Rafalski, J. A., & Tingey, S. V. (1990). DNA polymorphism amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Research, 18, 6531–6535.
Sarla, N., Bobba, S., & Siddiq, E. A. (2003). ISSR and SSR markers based on AG and GA repeats delineate geographically diverse Oryza nivara accessions and reveal rare alleles. Current Science, 84, 683–690.
Rohlf, F. J. (1993). NT-SYS-pc: numerical taxonomy and multivariate analysis system, version 2.11W. Setauket: Exteer software.
Esselman, E. J., Li, J. Q., Crawford, D., Winduss, J. L., & Wolfe, A. D. (1999). Clonal diversity in the rare Calamagrostis porter ssp. Insperata (Poaceae): comparative results for allozymes and random amplified polymorphic DNA (RAPD) and inter-simple sequence repeat (ISSR) markers. Molecular Ecology, 8, 443–451.
Li, Z., Liu, X., Gituru, R. W., Juntawong, N., Zhou, M., & Chen, L. (2010). Genetic diversity and classification of Nelumbo germplasm of different origins by RAPD and ISSR analysis. Scientia Horticulturae, 125, 724–732.
Nagaoka, T., & Ogihara, Y. (1997). Applicability of inter-simple sequence repeat polymorphisms in wheat for use as DNA markers in comparison to RFLP and RAPD markers. Theoretical and Applied Genetics, 94, 597–602.
Ajibade, S. R., Weeden, N. F., & Chite, S. M. (2000). Inter simple sequence repeat analysis of genetic relationships in the genus Vigna. Euphytica, 111, 47–55.
Yee, E., Kidwell, K. K., Sills, G. R., & Lumpkin, T. A. (1999). Diversity among selected Vigna angularis (Azuki) accessions on the basis of RAPD and AFLP markers. Crop Science, 39(268), 275.
Dwivedi, S. L., Gurtu, S., Chandra, S., Yuejin, W., & Nigam, S. N. (2001). Assessment of genetic diversity among selected groundnut germplasm I: RAPD analysis. Plant Breeding, 120(345), 349.
Acknowledgments
The authors are thankful to different farmers of Bihar and Research Centre Makhana, Darbhanga, for providing the diverse germplasms of E. ferox. This work is supported by a financial grant (Grant No.: SB/YS/LS-136/2013) from DST-SERB, New Delhi, India.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kumar, H., Priya, P., Singh, N. et al. RAPD and ISSR Marker-Based Comparative Evaluation of Genetic Diversity Among Indian Germplasms of Euryale ferox: an Aquatic Food Plant. Appl Biochem Biotechnol 180, 1345–1360 (2016). https://doi.org/10.1007/s12010-016-2171-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2171-z