Abstract
Agrobacterium-mediated sorghum transformation frequency has been enhanced significantly via medium optimization using immature embryos from sorghum variety TX430 as the target tissue. The new transformation protocol includes the addition of elevated copper sulfate and 6-benzylaminopurine in the resting and selection media. Using Agrobacterium strain LBA4404, the transformation frequency reached over 10% using either of two different selection marker genes, moPAT or PMI, and any of three different vectors in large-scale transformation experiments. With Agrobacterium strain AGL1, the transformation frequencies were as high as 33%. Using quantitative PCR analyses of 1,182 T0 transgenic plants representing 675 independent transgenic events, data was collected for T-DNA copy number, intact or truncated T-DNA integration, and vector backbone integration into the sorghum genome. A comparison of the transformation frequencies and molecular data characterizing T-DNA integration patterns in the transgenic plants derived from LBA4404 versus AGL1 transformation revealed that twice as many transgenic high-quality events were generated when AGL1 was used compared to LBA4404. This is the first report providing molecular data for T-DNA integration patterns in a large number of independent transgenic plants in sorghum.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sorghum (Sorghum bicolor L.) is the third most important cereal crop grown in the US and the fifth most important cereal crop grown in the world (US Grains Council). In 2012/2013, the total production of sorghum was 59.3 million metric tons worldwide (USDA Grain: World Markets and Trade, Feb. 2012/2013). Sorghum is used as the primary staple food for over 500 million people in the world, mainly in African and Asian countries. Some sorghum varieties are rich in antioxidants and all sorghum grain is gluten-free and thus provides an attractive grain replacement for celiac disease sufferers. Additionally, because this drought-resistant crop addresses global climate change and limited water resource issues, and because of increased interest in using sorghum for renewable fuel production, sorghum is important to both agriculture and industry.
Since the first successful Agrobacterium-mediated sorghum transformation protocol was published by our group in 2000 (Zhao et al. 2000), modifications of Agrobacterium-mediated transformation protocols have been reported (Jeoung et al. 2002; Carvalho et al. 2004; Gao et al. 2005a, b; Howe et al. 2006; Nguyen et al. 2007; Lu et al. 2009; Pandey et al. 2010; Shridhar et al. 2010). In these reports, based on small-scale transformation experiments, the highest transformation frequencies reported were 4.5% (Howe et al. 2006) and 4.28% (Shridhar et al. 2010). Alterations in parameters such as temperature and centrifugal force were investigated for their effects on transformation of immature embryos of rice and maize (Hiei et al. 2006) or of banana suspension cells (Khanna et al. 2004). Gurel et al. (2009) reported that pretreatment of immature embryos at 45°C for 3 min and cooling to 25°C before Agrobacterium infection resulted in an 8.3% transformation frequency of the sorghum variety P898012. Incremental improvements to the process have continued, and Liu and Godwin (2012) reported a 20.7% transformation frequency using microprojectile bombardment of the sorghum variety TX430. Unfortunately, despite production of transgenic sorghum with improved agronomic adaptability (Zhu et al. 1998; Krishnaveni et al. 2001; Girijashankar et al. 2005; Chandrashekar and Satyanarayana 2006; Kosambo-Ayoo et al. 2011) and nutrition (Zhao et al. 2003; da Silva et al. 2011), sorghum transformation is still not a robust, widely applicable technology (Shrawat and Lörz 2006; Pandey et al. 2010).
Recently, we realized that improved media formulations could play a substantial role in improving sorghum transformation, on a scale similar to what has been observed in response to increased levels of copper and 6-benzylaminopurine (BAP) in the medium used for barley transformation (Cho et al. 1998; Bregitzer et al. 2000).
Here, we report an optimized protocol for Agrobacterium-mediated sorghum transformation and the application of this modified protocol to large-scale sorghum transformation experiments. Using Agrobacterium strain LBA4404 and sorghum variety TX430, we generated 848 independent transgenic callus events and more than 1,300 transgenic T0 plants, utilizing approximately 7,000 immature embryos as transformation explant targets. The transformation frequency ranged from 8 to 13.4% among experiments using three different vectors. Furthermore, with this modified protocol, the transformation frequency increased to 33% by using the Agrobacterium strain AGL1. We also report the results of molecular analyses of 1,182 transgenic plants by quantitative PCR (Q-PCR). These transgenic plants represented 675 independent transgenic callus events derived from both AGL1 and LBA4404 transformation, with multiple plants being regenerated for many of the independent events. The molecular data for these transgenic plants included insert copy number, frequencies of intact or truncated T-DNA insertions, and frequencies of Agrobacterium vector backbone integration into the sorghum genome.
Materials and Methods
Plant material and transformation process.
TX430, a non-tannin sorghum variety, was used in this study. All sorghum embryo donor plants and transgenic plants were grown in a greenhouse located in Johnston, Iowa. Greenhouse temperatures averaged 29°C during the day and 20°C at night with a 12 h d/night photoperiod and supplemental lighting was provided by a 3:1 ratio of metal halide (1,000 W) and high-pressure sodium (1,000 W) lamps. The components of the media used in this study are listed in Table 1. The Escherichia coli phosphomannose isomerase (PMI) gene (Miles and Guest 1984), which catalyzes the reversible isomerization of mannose-6-phosphate to fructose-6-phosphate (serving as a precursor for the glycolytic pathway and enabling growth on mannose-containing media) and a codon-modified phosphinothricin acetyltransferase (moPAT) gene, which confers resistance to the herbicide glufosinate-ammonium (US patent no. 6,096,947), were used as the selectable markers. The baseline transformation protocol is described in detail as “treatment C” in Zhao et al. (2000). Briefly, freshly harvested sorghum immature grains were sterilized with 50% bleach and 0.1% Tween-20 for 30 min under vacuum and then rinsed with sterile water three times. The embryos were subjected to the following five sequential steps: (1) Agrobacterium infection: embryos were incubated in an Agrobacterium suspension (OD = 0.7 at 550 nm) with PHI-I medium for 5 min; (2) co-cultivation: embryos were cultured on PHI-T medium following infection for 3 d at 25°C in the dark; (3) resting: embryos were cultured on PHI-T medium plus 100 mg/l carbenicillin for 4 d at 28°C in the dark; (4) selection: embryos were cultured on PHI-U medium for 2 wk, followed by culture on PHI-V medium for the remainder of the selection process at 28°C in the dark, using subculture intervals of 2–3 wk; (5) regeneration: callus was cultured on PHI-X medium for 2–3 wk in the dark to stimulate shoot development, followed by culture for 1 wk under conditions of 16 h light (40–120 μE m−2 s−1) and 8 h dark at 25°C, and a final subculture on PHI-Z medium for 2–3 wk under lights (16 h, 40–120 μE m−2 s−1) to stimulate root growth. Regenerated plantlets were transplanted into soil and grown in the greenhouse (Zhao et al. 2000). T0 plants were self-pollinated to produce T1 progeny for further analysis. A new optimized protocol is described in the Results section of this manuscript.
Agrobacterium strains and vectors.
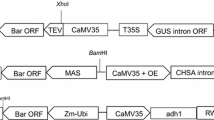
Agrobacterium tumefaciens strains LBA4404 and AGL1 (Lazo et al. 1991) were used in this study. Vectors pSB1 and pSB11 (Komari 1990; Komari et al. 1996) were used to construct the super-binary vectors PHP149, PHP166, and PHP32269 (Fig. 1). PHP149 contained three gene cassettes as follows: (1) the maize CZ19B1 promoter driving four inverted repeats for co-suppression of sorghum gamma-kafirin-1, gamma-kafirin-2, delta-kafirin-2, and lysine alpha-ketogluterate reductase; (2) the maize UBI promoter and UBI intron driving DsRed (Baird et al. 2000), which encodes a 28-kDa red fluorescent protein that serves as a visible marker; and (3) the maize UBI promoter and UBI intron driving PMI. PHP166 contained five gene cassettes as follows: (1) the maize CZ19B1 promoter driving inverted repeats of sorghum alpha-kafirin-A1 and gamma-kafirin-1 gene; (2) the maize oleosin promoter driving an inverted repeat of sorghum myo-inositol kinase (Shi et al. 2005); (3) the sorghum alpha-kafirin promoter driving maize phytoene synthase (Psy-1) (Buckner et al. 1996); (4) the sorghum beta-kafirin promoter driving Erwinia uredovora carotene desaturase (CrtI) (Ye et al. 2000); and (5) the maize UBI promoter and UBI intron driving PMI. PHP32269 contained two gene cassettes as follows: (1) the maize UBI promoter and UBI intron driving moPAT and yellow fluorescent protein (YFP, Nagai et al. 2002) and (2) the maize UBI promoter and UBI intron driving PMI.
The gene cassettes in the T-DNAs of PHP149, PHP166, and PHP32269. (A) T-DNA of PHP149 (11.8 kb) containing sequences for co-suppression of sorghum gamma-kafirin-1, gamma-kafirin-2, delta-kafirin-2, and sorghum LKR, and for expression of Ds-Red and PMI. (B) T-DNA of PHP166 (16.3 kb) containing sequences for co-suppression of sorghum gamma-kafirin-1, alpha-kafirin-A1, and sorghum MIK, and for expression of maize PSY-1, CRT-I, and PMI. The three regions for Q-PCR assays indicated with the bars underneath the vector map. (C) T-DNA of PHP32269 (7.9 kb) containing sequences for expression of PMI, the fusion protein gene moPAT, and YFP. The two regions for Q-PCR assays indicated with the bars underneath the vector map.
Quantitative PCR analysis of transgenic plants.
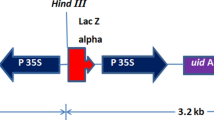
Q-PCR (Yang et al. 2009) was used to estimate the copy number of the transgenes to determine if T-DNA integrations were intact or truncated, and to screen for the presence of Agrobacterium binary vector backbone integration. Genomic DNA was extracted from a single piece (200 ng) of fresh leaf tissue from each plant (Truett et al. 2000). The non-transgenic sorghum line TX430 was used as the negative control and transgenic sorghum plants known to have single-copy transgenes were used as positive controls. Quantification was based on detection of amplification of gene sequences using gene-specific forward and reverse primers along with the corresponding gene-specific FAM/Vic-based MGB fluorogenic probes (Applied Biosystems, Foster City, CA). The 2−ΔΔCT method (Livak and Schmittgen 2001; ABI’s user bulletin #2, www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_040980.pdf) was used to estimate the copy number. For plants transformed with PHP166, three regions of the T-DNA (PMI, PSY-1, and CZ19B1 terminator (Fig. 1b ) were screened. Plants with single-copy scores for all of three sequences were classified as having intact single-copy T-DNA integrations. Plants that had single-copy scores for only one or two of the three sequences were classified as truncated single-copy events. Plants with multiple-copy scores for any of the three sequences were classified as having multiple-copy integrations, which could either be intact or truncated. Plants lacking all of these three sequences were classified as escapes. Plants were derived from PHP32269, primer sets that amplified segments of either the PMI or YFP; (Fig. 1c ). Detection of Agrobacterium vector backbone integrations derived from PHP166 or PHP32269 was based on screening for sequences from five regions outside of the T-DNA (near RB, virG, SPC, Tet, and near LB; Fig. 2; table of primer sequences available in supplemental data). Plants with negative Q-PCR signals for all five regions were considered as backbone-negative. Otherwise, they were classified as backbone-positive. Plants with intact single-copy T-DNA integrations without vector backbone were defined as quality events (QEs).
Statistics.
The chi-square test was used to analyze the segregation ratios of DsRed (+): DsRed (−) T1 plants transformed with PHP149 (Table 2). For each event, at least 100 T1 seeds (except for two events) were screened for DsRed expression, (excitation filter ET545/30 nm, emission filter ET620/60 nm). Chi-square values greater than 3.84 indicated that the observed ratio was different from the expected ratio for independent segregation of a single locus (3:1, P = 0.05). The PROC GLIMMIX procedure in SAS v.9.2 was used for data analysis. Transformation efficiency (stable events/embryos infected) was considered the response variable. A binomial distribution was assumed and the link function was logit. The estimation method was “LAPLACE.” For analysis of plants transformed with PHP166, the individual researcher (transformer) was considered as a fixed effect. Overdispersion was addressed by adding a multiplicative overdispersion factor in the model. For comparison of selection markers, the selection marker was considered as a fixed effect. For comparison of Agrobacterium strains, the strain was considered as a fixed effect. Because experiments were performed side by side across numerous replicates, experiment was considered as a random effect.
Results
Enhancement of transformation frequency via media modification.
Because TX430 is a non-tannin sorghum line that produces low levels of phenolics during tissue culture, polyvinylpolypyrrolidone was not added to any medium used in the transformation process. Agrobacterium strain LBA4404 containing the vector PHP149 was used in these experiments because the visible marker DsRed aided in identification of transgenic callus sectors at different transformation steps and in regenerated plants. Through a series of medium modifications and tests (data not shown), use of medium DBC3 (Cho et al. 1998; Table 1) significantly improved transformation efficiency when it was used in both the resting and selection steps, but had a negative impact when used during co-cultivation.
Comparison of DBC3-based media to PHI-T during the resting and PHI-UM during the selection steps demonstrated the benefit of using DBC3-based medium. Transformation frequencies were calculated based on the number of recovered T0 transformation events capable of growth on mannose-containing medium (indicating expression of PMI) and of expression of DsRed in shoots, roots, leaves, and pollen (Fig. 3) relative to the number of starting immature embryos used for transformation. Use of the baseline protocol resulted in the recovery of T0 plants from two independent transgenic events derived from 362 immature embryos, for a transformation frequency of 0.6%. By comparison, use of the DBC3 media for the resting and selection steps resulted in the recovery of T0 plants from 29 independent transgenic events derived from 361 immature embryos, for a transformation frequency of 8%. From these 29 events, 170 T0 plants (4–6 T0 plants per event) were grown and self-pollinated in the greenhouse, and 161 plants (95%) produced T1 seeds. Based on DsRed expression (Fig. 3e ), a 3 red/1 non-red ratio was observed for 18 events (66.7%, Table 2). A higher ratio than expected was observed for seven events (25.9%), and a lower ratio than expected was observed for two events (7.4%). From the two transgenic events recovered using the treatment C protocol, 11 of 12 T0 plants (6 from each event) grown in the greenhouse produced T1 seed. The expected ratio of 3 red/1 non-red T1 was observed for both events.
Transgenic callus, shoots, roots, leaf, pollen, and seeds expressing DsRed derived from PHP149 viewed using fluorescence microscopy. (a) Transgenic callus expressing DsRed, (b) transgenic shoots and roots expressing DsRed, (c) transgenic leaf expressing DsRed, (d) pollen grains from a T0 transgenic plant with or without expression of DsRed, (e) T1 seeds from a T0 transgenic plant with or without expression of DsRed, (f) a part of a T0 transgenic panicle showing red (transgenic) and non-red (non-transgenic) seeds.
Large-scale sorghum transformation with the modified protocol.
To validate the utility of this new protocol, we applied it to a large-scale sorghum transformation experiment using Agrobacterium strain LBA4404 and vector PHP166. This vector was designed to improve the levels of pro-vitamin A, mineral bioavailability, protein quality, and protein digestibility in sorghum seed.
A total of 6,077 immature embryos were treated with LBA4404/PHP166 over the course of 23 experiments, producing 755 independent putative transgenic callus events that regenerated into plants. Transformation frequencies, based on the ability to grow on mannose-containing medium, were calculated and ranged from 2.3 to 26.3% among experiments, with a mean of 12.4%. Transformation frequencies were below 10% in nine experiments, between 10 and 20% in ten experiments, and higher than 20% in four experiments. Mean transformation frequencies achieved by each of three individual transformers achieved average transformation frequencies of 9.7, 13, and 15%, which were confirmed to be statistically similar (P = 0.49). Not all events were processed for molecular analysis. Instead, two T0 plants were regenerated from a subset of 507 callus events, and all of these 1,014 T0 plants were moved to the greenhouse for later sampling and further analysis (Fig. 4). Leaf samples were collected from each T0 plant, and extracted DNA was analyzed using Q-PCR to (1) detect stable integration of the T-DNA into the sorghum genome, (2) determine copy number of T-DNA integrations, (3) determine whether integrated T-DNA was intact or truncated, and (4) detect the presence of plasmid backbone.
These analyses showed that all 1,014 T0 plants were transgenic, and that the transformation frequencies calculated based on growth on mannose-containing medium were correct. The plants were divided into three categories based on Q-PCR data (Table 3). In the first category, 382 T0 plants (37.7%) contained single-copy intact integrations of the T-DNA. In the second category, 269 plants (26.5%) contained single-copy but truncated integrations of the T-DNA. In the final category, 363 plants (35.8%) contained multiple copies of the T-DNA (either intact or truncated).
The 382 T0 plants containing single-copy T-DNA inserts (category 1) were further analyzed to detect unintended integration of the vector backbone. No backbone was detected in 359 (94%) of these 382 T0 plants. Based on this analysis, we designated the transgenic plants or events with intact single copy of T-DNA integration and no vector backbone as QEs. The 359 QEs were grown to maturity in the greenhouse and self-pollinated (Fig. 4), while all remaining non-QE events (with vector backbone or from groups 2 and 3) were discarded. From the 359 self-pollinated T0 plants, 343 (95.5%) produced T1 seeds. These 343 plants were originally regenerated from 172 callus events. Thus, from a population of 507 transgenic events analyzed using Q-PCR, 33.9% (172/507) ended up being “quality” events that transmitted the single-copy transgenic locus to progeny.
Comparison of moPAT and PMI as selection markers in sorghum line TX430.
Vector PHP32269 in Agrobacterium LBA4404 contained two selectable marker genes, moPAT, and PMI (Fig. 1), permitting the direct comparison of these markers during the transformation process. For these experiments, we used the DBC3 base medium containing either 3 mg/l bialaphos (for the moPAT gene) or 12.5 g/l mannose for the PMI gene. For 6 experiments that used moPAT-based selection, 43 independent transgenic events produced T0 plants that were obtained from 320 embryos for an average transformation frequency of 13.4% (range of 8–17.5% among experiments). For 5 experiments that used PMI-based selection, 21 independent transgenic events that produced T0 plants were obtained from 216 embryos, for an average transformation frequency of 9.7% (range of 6.1–14.6% among experiments). The transgenic nature of these T0 plants was confirmed by YFP expression. Statistical analysis indicated no significant difference between the transformation frequencies obtained using these two selectable markers (P = 0.19).
Comparison of Agrobacterium strain AGL1 and LBA4404 in sorghum line TX430.
These experiments used vector PHP32269, and PMI was used as the selectable marker. Over 5 experiments, 286 immature embryos were infected using AGL1, producing 95 independent transgenic events for an average transformation frequency of 33.2% (range of 18.1 to 44.1% among experiments). In four of five experiments, the transformation frequencies were over 28.9%, and in two, transformation frequencies were over 41.2%. For LBA4404, 216 embryos were initially infected, producing 21 independent transgenic events for an average transformation frequency of 9.7%, (range of 6.1 to 14.6%). Overall, the transformation frequency using AGL1 was 3.4-fold higher than that obtained with LBA4404 (P = 1.83 × 10−9).
T-DNA integrations in T0 plants from 113 AGL1-derived events (95 events from the above experiments and 18 events from experiments not shown) and 55 LBA4404-derived transformation events (21 events from the above experiments and 34 events from experiments not shown) were analyzed by Q-PCR (Table 4). Of the 113 plants from AGL1, 46 (40.7%) harbored intact single-copy T-DNA integrations, while 67 (59.3%) contained either multiple-copy integrations or truncated single-copy integrations. Of the 46 AGL1-derived plants containing intact single-copy integrations, 34 (74%) did not contain vector backbone. Thus, the overall frequency of transgenic events meeting the criteria for QEs derived from AGL1 was 30% (34/113). Of the 55 LBA4404-derived plants, 28 (51%) contained intact single copy T-DNA integrations, while 27 (49%) contained either multi-copy integrations or truncated single-copy integrations. Of the 28 plants harboring intact single-copy integrations, 27 (96.4%) contained no vector backbone and the overall QE frequency was 49% (27/55). In summary, a higher transformation frequency (33.2%) but a lower frequency of QE production (30%) was observed when AGL1 was used than when LBA4404 was used (9.7% transformation frequency and 49% QE production). The overall transformation frequency obtained when AGL1 was used (combining transformation frequency and QE frequency), however, was still much higher than when LBA4404 was used. One QE could be obtained from 10 infected embryos using AGL1, whereas 21 infected embryos were required to obtain 1 QE when using LBA4404.
New sorghum transformation protocol.
Through a series of medium modifications and transformation evaluations described above, we have established a new sorghum transformation protocol
-
1.
Infect freshly isolated embryos with Agrobacterium (either AGL1 or LBA4404) suspension (OD = 0.7 at 550 nm) in PHI-I medium for 5 min.
-
2.
Culture the infected embryos on PHI-T medium for 4–7 d at 25°C in the dark for co-cultivation.
-
3.
Culture the infected embryos on DBC3 medium plus 100 mg/l carbenicillin (if using LBA4404) or 100 mg/l cefotaxime and 150 mg/l timentin (if using AGL1) for 7–10 d at 28°C in the dark for resting.
-
4.
Start selection on the 14th day from infection on DBC3 (plus 100 mg/l carbenicillin, if using Agrobacterium strain LBA4404; or plus100 mg/l cefotaxime and 150 mg/l timentin, if using Agrobacterium strain AGL1).
-
(a)
Add 12.5 g/l mannose and reduce the maltose concentration to 5 g/l if using PMI as the selection marker, or add 3 mg/l bialaphos if moPAT is used as the selection marker
-
(b)
Subculture on fresh medium every 2–3 wk for a total duration of approximately 2.5–3 mo
-
(a)
-
5.
Initiate shoot development on PHI-XM medium for 2–3 wk in the dark, and then for 1 wk under light.
-
6.
Initiate root development on PHI-Z medium for 2–3 wk in the light.
-
7.
Transplant the regenerated plantlets into soil for growth to maturity and seed production.
Discussion
Sorghum, a drought-tolerant crop, has attracted increased attention in recent years, with an interest in producing genetically modified sorghum. However, the low efficiency of sorghum transformation has continued to be a bottleneck for both academics and industry. Here, we report an optimized sorghum transformation protocol, relative to the protocol first described by Zhao et al. (2000) and we present data derived from experiments using the variety TX430 to demonstrate this improvement. Similar to results first reported in barley (Cho et al. 1998), we observed that increasing the concentration of copper in the form of cupric sulfate (from 0.1 to 5 μM) and the addition of the cytokinin BAP in the resting and selection stages of the process played a critical role in enhancing sorghum transformation by promoting the generation of high-quality, fast-growing, and regenerable transgenic callus. We also evaluated a range of copper concentrations and found that copper levels from 1 to 50 μM in the resting and selection media did not alter the sorghum transformation frequency (data not shown). However, if elevated copper and additional BAP were added to the co-cultivation medium (along with resting and selection media), the transformation frequencies were even lower than that described in our baseline treatment C medium (Zhao et al. 2000). Therefore, low copper and no BAP during co-cultivation, followed by elevated copper and BAP during the resting and selection stages are important components of this new protocol. The reproducibility of this protocol was demonstrated by the consistently high transformation frequencies generated from large numbers of immature embryos across multiple experiments. With this optimized protocol, the transformation frequencies using Agrobacterium strain LBA4404 and two selection marker genes, moPAT and PMI, across three different vectors were very consistent: 8% for PHP149 with PMI selection, 12.4% for PHP166 with PMI selection, and 13.4 (moPAT selection) or 9.7% (PMI selection) with PHP32269. Using this optimized protocol with Agrobacterium strain AGL1, the transformation frequency increased to levels as high as 33.2%, substantially higher than previously reported efficiencies in sorghum.
Molecular analyses of sorghum transgenic plants collected from 1,182 T0 plants representing 675 independent transgenic events. All of these molecular data were based on Q-PCR analysis. A major advantage to using multiplex Q-PCR (to cover multiple genes within the expression cassette) is the high degree of confidence this data provides, in terms of verifying a transgenic event. We covered three regions on vector PHP166, two regions on vector PHP32269, and five regions of vector backbone for Q-PCR analysis. This large-scale dataset provided a clear picture of T-DNA integration patterns in the sorghum genome, demonstrating that the frequency of producing “quality” (intact single copy of T-DNA without vector backbone presence) transgenic sorghum events is impacted by at least two factors—the Agrobacterium strain and the size of T-DNA. Based on data from side-by-side transformation comparisons, AGL1 produced a higher transformation frequency but lower quality event frequency than LBA4404. When LBA4404 was used in sorghum transformation, the quality event frequency was 49% for vector PHP32269 and 35.4% for vector PHP166, in which the size of the T-DNA was 7.9 and 16.3 kb, respectively. Whether the increased size of the T-DNA used in Agrobacterium-mediated transformation consistently reduces quality event frequency needs more data for elucidation (Song et al. 2003).
Q-PCR as a method to estimate copy number of transgenes in transgenic plants has been investigated by several groups. Song et al. (2002) compared transgene copy number estimated by Q-PCR and by Southern analysis in WHISKERSTM-derived transgenic maize callus and plants containing either the A0 gene or the IMT gene. A strong correlation between the copy number as determined by Q-PCR and by Southern analysis was demonstrated and Q-PCR seemed to provide good estimations of transgene copy number in transgenic maize callus and plants. Chu et al. (2013) reported a correlation coefficient of 0.95 between transgene copy number estimated by Q-PCR versus Southern blot analysis in 30 independent transgenic peanut lines. Others have reported similar results (Ingham et al. 2001; Song et al. 2002; Shou et al. 2004). Bubner and Baldwin (2004) reviewed the function of conventional-PCR, competitive-PCR, and quantitative-PCR and pointed out that Q-PCR was able to detect a twofold difference when used to estimate the transgene copy number in plants. It is particularly useful as a preliminary screening tool for estimating copy numbers of a large number of transformants. Bubner and Baldwin also commented that Q-PCR is not fail-safe, and should be viewed as complementary to, rather than as a replacement of, other methods such as Southern analysis. Based on these reports and the data presented in this study, we feel that Q-PCR can be used with confidence as a starting point to sort transgenic events for product development. More accurate analyses such as Southern analysis, flanking sequence analysis, and sequencing of the T-DNA insertions, usually performed on selected plants or events at the later stage of product development, will reveal additional detailed molecular information on the nature of T-DNA integrations.
Conclusion
Both transformation frequency and quality event frequency were used to provide a benchmark for evaluation of transformation protocols. Based on our new protocol, transformation of 100 sorghum immature embryos with LBA4404 will generate 4 to 5 independent quality events. With AGL1, 10 independent quality events can be produced. This optimized sorghum transformation protocol and the molecular analysis of the transgenic plants should help sorghum biotechnology progress, and allow greater progress to be made for both basic and applied research with this important crop plant.
References
Baird G. S.; Zacharias D. A.; Tsien R. Y. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. U. S. A. 97: 11984–11989; 2000.
Bregitzer P.; Campbell R. D.; Dahleen L. S.; Lemaux P. G.; Cho M.-J. Development of transformation systems for elite barley cultivars. Barley Genet. Newsl. 30: 10; 2000.
Bubner B.; Baldwin I. T. Use of real-time PCR for determining copy number and zygosity in transgenic plants. Plant Cell Rep. 23: 263–271; 2004.
Buckner B.; San-Miguel P.; Bennetzen J. L. The y1 gene of maize codes for phytoene synthase. Genetics 143: 479–488; 1996.
Carvalho C. H. S.; Zehr U. B.; Gunaratna N.; Anderson J.; Kononowicz H. H.; Hodges T. K.; Axtell J. D. Agrobacterium-mediated transformation of sorghum: factors that affect transformation efficiency. Genet. Mol. Biol 27: 259–269; 2004.
Chandrashekar A.; Satyanarayana K. V. Disease and pest resistance in grains of sorghum and millets. J. Cereal Sci 44: 287–304; 2006.
Cho M.-J.; Jiang W.; Lemaux P. G. Transformation of recalcitrant barley cultivars through improvement of regenerability and decreased albinism. Plant Sci 138: 229–244; 1998.
Chu Y.; Bhattacharya A.; Wu C.; Knoll J. E.; Ozias-Akins P. Improvement of peanut (Arachis hypogaea L.) transformation efficiency and determination of transgene copy number by relative quantitative real-time PCR. In Vitro Cell Dev. Biol–Plant 49: 266–275; 2013.
da Silva L. S.; Jung R.; Zhao Z. Y.; Glassman K.; Taylor J.; Taylor J. R. N. Effect of suppressing the synthesis of different kafirin sub-classes on grain endosperm texture, protein body structure, and protein nutritional quality in improved sorghum lines. J Cereal Sci 54: 160–167; 2011.
Gao Z.; Jayaraj J.; Muthukrishnan S.; Claflin L.; Liang G. H. Efficient genetic transformation of sorghum using a visual screening marker. Genome 48: 321–333; 2005a.
Gao Z.; Xie X.; Ling Y.; Muthukrishnan S.; Liang G. H. Agrobacterium tumefaciens-mediated sorghum transformation using a mannose selection system. Plant Biotechnol. J 3: 591–599; 2005b.
Girijashankar V.; Sharma H. C.; Sharma K. K.; Swathisree V.; Prasad L. S.; Bhat B. V.; Royer M.; Secundo B. S.; Narasu M. L.; Altosaar I.; Seetharama N. Development of transgenic sorghum for insect resistance against the spotted stem borer (Chilo partellus). Plant Cell Rep. 24: 513–522; 2005.
Gurel S.; Gurel E.; Kaur R.; Wong J.; Meng L.; Tan H.-Q.; Lemaux P. G. Efficient, reproducible Agrobacterium-mediated transformation of sorghum using heat treatment of immature embryos. Plant Cell Rep. 28: 429–444; 2009.
Hiei Y.; Ishida Y.; Kasaoka K.; Komari T. Improved frequency of transformation in rice and maize by treatment of immature embryos with centrifugation and heat prior to infection with Agrobacterium tumefaciens. Plant Cell, Tissue Organ Cult. 87: 233–243; 2006.
Howe A.; Sato S.; Dweikat I.; Fromm M.; Clemente T. Rapid and reproducible Agrobacterium-mediated transformation of sorghum. Plant Cell Rep. 25: 784–791; 2006.
Ingham D. J.; Beer S.; Money S.; Hansen G. Quantitative real-time PCR assay for determining transgene copy number in transformed plants. Biotechniques 31: 132–140; 2001.
Jeoung J. M.; Krishnaveni S.; Muthukrishnan S.; Trick H. N.; Liang G. H. Optimization of sorghum transformation parameters using genes for green fluorescent protein and β-glucuronidase as visual markers. Hereditas 137: 20–28; 2002.
Khanna H.; Becker D.; Kleidon J.; Dale J. Centrifugation assisted Agrobacterium tumefaciens-mediated transformation (CAAT) of embryogenic cell suspensions of banana (Musa spp. Cavendish AAA and Lady finger AAB). Mol. Breeding 14: 239–252; 2004.
Komari T. Transformation of cultured cells of Chenopodium quinoa by binary vectors that carry a fragment of DNA from the virulence region of pTiBo542. Plant Cell Rep. 9: 303–306; 1990.
Komari T.; Hiei Y.; Saito Y.; Murai N.; Kumashiro T. Vectors carrying two separate T-DNAs for co-transformation of higher plants mediated by Agrobacterium tumefaciens and segregation of transformants free from selection markers. Plant J. 10: 165–174; 1996.
Kosambo-Ayoo L. M.; Bader M.; Loerz H.; Becker D. Transgenic sorghum Sorghum bicolor L. Moench) developed by transformation with chitinase and chitosanase genes from Trichoderma harzianum expresses tolerance to anthracnose. Afr. J. Biotechnol 10: 3659–3670; 2011.
Krishnaveni S.; Jeoung J. M.; Muthukrishnan S.; Liang G. H. Transgenic sorghum plants constitutively expressing a rice chitinase gene show improved resistance to stalk rot. J. Genet. Breed 55: 151–158; 2001.
Lazo G. R.; Stein P. A.; Ludwig R. A. A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology 9: 963–967; 1991.
Liu G.; Ian G. D. Highly efficient sorghum transformation. Plant Cell Rep. 31: 999–1007; 2012.
Livak K. J.; Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408; 2001.
Lu L.; Wu X.; Yin X.; Morranf J.; Chen X.; Folk W. R.; Zhang Z. J. Development of marker-free transgenic sorghum [Sorghum bicolor (L.) Moench] using standard binary vectors with bar as a selectable marker. Plant Cell, Tissue Organ Cult. 99: 97–108; 2009.
Miles J. S.; Guest J. R. Nucleotide sequence and transcriptional start point of the phosphomannose isomerase (manA) gene of Escherichia coli. Gene 32: 41–48; 1984.
Nagai T.; Ibata K.; Park E. S.; Kubota M.; Mikoshiba K.; Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell biological applications. Nat. Biotechnol. 20: 87–90; 2002.
Nguyen T. V.; Thu T. T.; Claeys M.; Angenon G. Agrobacterium-mediated transformation of sorghum (Sorghum bicolor (L.) Moench) using an improved in vitro regeneration system. Plant Cell, Tissue Organ Cult. 91: 155–164; 2007.
Pandey A. K.; Bhat B. V.; Balakrishna D.; Seetharama N. Genetic transformation of sorghum (Sorghum bicolor (L.), Moench). Int. J. Biotechnol. Biochem 6: 45–53; 2010.
Shi J.; Wang H.; Hazebroek J.; Ertl D. S.; Harp T. The maize low-phytic acid 3 encodes a myo-inositol kinase that plays a role in phytic acid biosynthesis in developing seeds. Plant J. 42: 708–719; 2005.
Shou H.; Frame B. R.; Whitham S. A.; Wang K. Assessment of transgenic maize events produced by particle bombardment or Agrobacterium-mediated transformation. Mol Breeding 13: 201–208; 2004.
Shrawat H. K.; Lörz H. Agrobacterium-mediated transformation of cereals: a promising approach crossing barriers. Plant Biotechnol. J 4: 575–603; 2006.
Shridhar J.; Bhat R. S.; Bhat S.; Kuruvinashetti M. S. Agrobacterium-mediated transformation studies in sorghum using an improved gfp reporter gene. J. SAT Agric. Res 8: 1–5; 2010.
Song J.; Bradeen J. M.; Naess S. K.; Helgeson J. P.; Jiang J. BIBAC and TAC clones containing potato genomic DNA fragments larger than 100 kb are not stable in Agrobacterium. Theor. Appl. Genet. 107: 958–964; 2003.
Song P.; Cai C. Q.; Skokut M.; Kosegi B. D.; Petolino J. F. Quantitative real-time PCR as a screening tool for estimating transgene copy number in WHISKERSTM-derived transgenic maize. Plant Cell Rep. 20: 948–954; 2002.
Truett G. E.; Heeger P.; Mynatt R. L.; Truett A. A.; Walker J. A.; Warman M. L. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 29: 52–54; 2000.
Yang M.; Djukanovic V.; Stagg J.; Lenders B.; Bidney D.; Falco S. C.; Lyznik L. A. Targeted mutagenesis in the progeny of maize transgenic plants. Plant Mol. Biol. 70: 669–679; 2009.
Ye X.; Al-Babili S.; Klo” Ti A.; Zhang J.; Lucca P.; Beyer P.; Potrykus I. Engineering the provitamin A (b-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287: 303–305; 2000.
Zhao Z. Y.; Cai T.; Tagliani L.; Miller M.; Wang N.; Pang H.; Rudert M.; Schroeder S.; Hondred D.; Seltzer J.; Pierce D. Agrobacterium-mediated sorghum transformation. Plant Mol. Biol. 44: 789–798; 2000.
Zhao Z. Y.; Glassman K.; Sewalt V.; Wang N.; Miller M.; Chang S.; Thompson T.; Catron S.; Wu E.; Bidney Y.; Jung R. Nutritionally improved transgenic sorghum. In: Vasil I. K. (ed) Plant biotechnology 2002 and beyond. Kluwer Academic Publishers, Dordrecht, pp 413–416; 2003.
Zhu H.; Muthukrishnan S.; Krishnaveni S.; Wilde G.; Jeoung J. M.; Liang G. H. Ballistic transformation of sorghum using a rice chitinase gene. J. Genet. Breed 52: 243–252; 1998.
Acknowledgments
This work was partially funded by the Bill and Melinda Gates Foundation for Global Challenge (GC) #9 project: “Nutritionally-enhanced Sorghum for the arid and semiarid tropical areas of Africa.” This manuscript was critically reviewed by Drs. Marc Albertsen, James Register, Todd Jones, and William Gordon-Kamm. Dr. Hua Mo provided the statistical analysis of the transformation data.
Author information
Authors and Affiliations
Corresponding author
Additional information
Editor: Lynn Dahleen
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 14.1 KB)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Wu, E., Lenderts, B., Glassman, K. et al. Optimized Agrobacterium-mediated sorghum transformation protocol and molecular data of transgenic sorghum plants. In Vitro Cell.Dev.Biol.-Plant 50, 9–18 (2014). https://doi.org/10.1007/s11627-013-9583-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11627-013-9583-z