Abstract
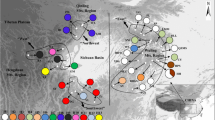
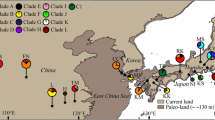
In this study, we investigated two chloroplast intergenic spacers (accD-psaI and trnL-trnF) for a total of 266 individuals over 22 populations of Pyrus pashia to detect the genetic diversity, genetic, and phylogeographic structure and provide needed information for the development of conservation strategies. Thirteen haplotypes (H1–H13) were recognized. A high level of total diversity was detected (H T = 0.746). Analysis of molecular variance indicated that the genetic variation mainly existed within populations, representing 59.61 % of the total variation. Genetic differentiation among populations was high (F st = 0.404). A Mantel test did not show a correlation between the genetic and geographic distances (r = 0.139, P = 0.09, 1,000 permutations), implying that geographic distance was not critical to gene flow among populations. A significantly higher N st than G st (N st = 0.420, G st = 0.402, P < 0.05) reflected the phylogeographic structure in P. pashia. While nested clade analyses of clade 2-2 showed that restricted gene flow existed among populations, clades 1-2 and 2-1, and the total cladogram exhibited contiguous range expansion events in P. pashia. Both sum of square deviations and the raggedness index failed to reject the sudden demographic expansion model. The overall population expansion of P. pashia was estimated to occur between 621,000 and 209,000 years ago.


Similar content being viewed by others
References
Avise JC (1994) Molecular markers, natural history, and evolution. Chapman & Hall, New York
Avise JC (2000) Phylogeography: the history and formation of species. Harvard University Press, Cambridge
Bailey LH (1917) Pyrus. In: Standard cyclopedia of horticulture, vol V. Macmillan, New York, pp 2865–2878
Bao L, Chen KS, Zhang D, Cao YF, Yamamoto T, Teng YW (2007) Genetic diversity and similarity of pear cultivars native to East Asia revealed by SSR (simple sequence repeat) markers. Genet Resour Crop Evol 54:959–971
Bao L, Chen KS, Zhang D, Li XG, Teng YW (2008) An assessment of genetic variability and relationships within Asian pears based on AFLP (amplified fragment length polymorphism) markers. Sci Hortic 116:374–380
Chiang TY, Chiang YC, Chen YJ, Chou CH, Havanond S, Hong TN, Huang S (2001) Phylogeography of Kandelia candel in East Asiatic mangroves based on nucleotide variation of chloroplast and mitochondrial DNAs. Mol Ecol 10:2697–2710
Clegg MT, Zurawski G (1992) Chloroplast DNA and the study of plant phylogeny: present status and future prospects. Chapman & Hall, New York
Clement M, Posda D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferblack from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0 An integrated software package for population genetics data analysis. Evol Bioinformatics Online 1:47–50
Ferris C, King RA, Vainola R, Hewitt GM (1998) Chloroplast DNA recognizes three refugia sources of European oaks and suggests independent eastern and western immigrations to Finland. Heredity 80:584–593
Gong X, LuanSS HKH, Hwang CC, Lin CJ, Chiang YC, Chiang TY (2011) Population structure of Nouelia insignis (Asteraceae), an endangered species in southwestern China, based on chloroplast DNA sequences: recent demographic shrinking. J Plant Res 124:221–230
Graur D, Li WH (1999) Fundamentals of molecular evolution. Sinauer Associates, Sunderland
Harpending HC (1994) Signature of ancient population-growth in a low-resolution mitochondrial-DNA mismatch distribution. Hum Biol 66:591–600
Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc 58:247–276
Hewitt GM (2001) Speciation, hybrid zones and phylogeography—or seeing genes in space and time. Mol Ecol 10:537–549
Hu CY, Zheng XY, Teng YW (2011) Characterization and phylogenetic utility of non-coding chloroplast regions trnL-trnF and accD-psaI in Pyrus. Acta Hortic Sina 38:2261–2272 (in Chinese)
Huang S, Chiang YC, Schaal BA, Chou CH, Chiang TY (2001) Organelle DNA phylogeography of Cycas taitungensis, a relict species in Taiwan. Mol Ecol 10:2669–2681
Iketani H, Manabe T, Matsuta N, Akihama T, Hayashi Y (1998) Incongruence between RFLPs of chloroplast DNA and morphological classification in east Asian pear (Pyrus spp.). Genet Resour Crop Evol 45:533–539
Katayama H, Tachibana M, Iketani H, Zhang SL, Uematsu C (2012) Phylogenetic utility of structural alterations found in the chloroplast genome of pear: hypervariable regions in a highly conserved genome. Tree Genet Genomes 8:313–326
Kimura T, Iketani H, Kotobuki K, Matsuta N, Ban Y, Hayashi T, Yamamoto T (2003) Genetic characterization of pear varieties revealed by chloroplast DNA sequences. J Hortic Sci Biotechnol 78:241–247
Li N, Fu L (1997) Notes on gymnosperms I. Taxonomic treatment of some Chinese conifers. Novon 7:261–264
Liu J, Zheng XY, Potter D, Hu CY, Teng YW (2012) Genetic diversity and population structure of Pyrus calleryana (Rosaceae) in Zhejiang province, China. Biochem Syst Ecol 45:69–78
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Newton AC, Allnutt AR, Gillies ACM, Lowe AJ, Ennos RA (1999) Molecular phylogeography, intraspecific variation and the conservation of tree species. Trends Ecol Evol 14:140–145
Petit RJ, Aguinagalde I, de Beaulieu JL, Bittkau C, Brewer S et al (2003) Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300:1563–1565
Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin GG (2005) Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol Ecol 14:689–701
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
Posada D, Crandall KA (2001) Intraspecific gene genealogies: trees grafting into networks. Trends Ecol Evol 16:37–45
Posada D, Crandall KA, Templeton AR (2000) GeoDis: a program for the cladistic nested analysis of geographical distribution of genetic haplotypes. Mol Ecol 9:487–488
Potter D, Eriksson T, Evans RC, Oh S, Smedmark JEE, Morgan DR, Kerr M, Robertson KR, Arsenault M, Dickinson TA, Campbell CS (2007) Phylogeny and classification of Rosaceae. Plant Syst Evol 266:5–43
Rozas J, Sanchez-Delbarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Rubtsov GA (1944) Geographic distribution of the genus Pyrus and trends and factors in its evolution. Am Soc Nat 78:358–366
Schaal BA, Hayworth DA, Olsen KM, Rauscher JT, Smith W (1998) Phylogeography studies in plants: problems and prospects. Mol Ecol 7:465–474
Small RL, Ryburn JA, Cronn RC, Seelanan T, Wendel JF (1998) The tortoise and the hare: choosing between noncoding plastome and nuclear ADH sequences for phylogeny reconstruction in recently diverged plant group. Am J Bot 85:1301–1345
Taberlet P, Fumagalli L, Wust-Saucy AG, Cosson JF (1998) Comparative phylogeography and postglacial colonisation routes in Europe. Mol Ecol 7:453–464
Templeton AR, Routman E, Phillips CA (1995) Separating population structure from population history: a cladistic analysis of the geographical distribution of mitochondrial DNA haplotypes in the tiger salamander, Ambystoma tigrinum. Genetics 140:767–782
Teng YW, Tanabe K, Tamura F, Itai A (2001) Genetic relationships of pear cultivars in Xinjiang, China as measublack by RAPD markers. J Hortic Sci Biotech 76:771–779
Teng YW, Tanabe K, Tamura F, Itai A (2002) Genetic relationship of Pyrus species and cultivars native to east Asia revealed by randomly amplified polymorphic DNA makers. J Am Soc Hortic Sci 127:262–270
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL X: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Wang ZJ, Guan KY (2011) Genetic structure and phylogeography of a relict tree fern, Sphaeropteris brunoniana (Cyatheaceae) from China and Laos inferred from cpDNA sequence variations: implication for conservation. J Syst Evol 49:71–79
Wolfe KH, Li WH, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondria, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA 84:9054–9058
Xue WQ, Zhang XS (2011) Geographic distribution of Coenosia Meigen (Diptera, Muscidae) of Yunnan, China, with descriptions of four new species. Dtsch Entomol Z 58:155–164
Yamamoto T, Kimura T, Sawamura Y, Kotobuki K, Ban Y, Hayashi T, Matsuta N (2001) SSRs isolated from apple can identify polymorphism and genetic diversity in pear. Theor Appl Genet 102:865–887
Yamamoto T, Kimura T, Sawamura Y, Manabe T, Kotobuki K, Hayashi T, Ban Y, Matsuta N (2002a) Simple sequence repeats for genetic analysis in pear. Euphytica 124:129–137
Yamamoto T, Kimura T, Sawamura Y, Manabe T, Kotobuki K, Hayashi T, Ban Y, Matsuta N (2002b) Genetic linkage maps constructed by using an interspecific cross between Japanese and European pears. Theor Appl Genet 106:9–18
Yao LH, Zheng XY, Cai DY, Gao Y, Wang K, Cao YF, Teng YW (2010) Exploitation of Malus EST-NSSRs and the utility in evaluation of genetic diversity in Malus and Pyrus. Genet Resour Crop Evol 57:841–851
Yu DJ, Lu LD, Gu CZ, Guan KJ, Jiang WF (1986) Flora Republicae Popularis Sinicae 38:367–370
Zhang Q, Chiang TY, George M, Liu JQ, Abbott RJ (2005) Phylogeography of the Qinghai-Tibetan Plateau endemic Juniperus przewalskii (Cupressaceae) inferred from chloroplast DNA sequence variation. Mol Ecol 14:3513–3524
Zheng XY, Hu CY, Spooner D, Liu J, Cao JS, Teng YW (2011) Molecular evolution of Adh and LEAFY and the phylogenetic utility of their introns in Pyrus (Rosaceae). BMC Evol Biol 11:255
Acknowledgments
We sincerely thank En-Sheng Dong form the Fruit and Vegetable Work Station of Weining County, Guizhou Province, China, for his help in sampling P. pashia populations in Guizhou Province. This project was supported by the National Natural Science Foundation of China (no. 30871690).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E. Dirlewanger
Rights and permissions
About this article
Cite this article
Liu, J., Sun, P., Zheng, X. et al. Genetic structure and phylogeography of Pyrus pashia L. (Rosaceae) in Yunnan Province, China, revealed by chloroplast DNA analyses. Tree Genetics & Genomes 9, 433–441 (2013). https://doi.org/10.1007/s11295-012-0564-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-012-0564-x