Abstract
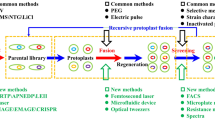
Genome shuffling was used to improve the thermotolerance of l-glutamic acid-producing strain Corynebacteria glutamicum. Five strains with subtle improvements in high temperature tolerance and productivity were selected by ultraviolet irradiation and diethyl sulfate mutagenesis. An improved strain (F343) was obtained by three rounds of genome shuffling of the five strains as mentioned above. The cell density of F343 was four times higher than that of ancestor strains after 24 h of cultivation at 44°C, and importantly, the yield of l-glutamic acid was increased by 1.8-times comparing with that of the ancestor strain at 38°C in a 5-L fermentor. With glucose supplement and two-stage pH control, the l-glutamate acid concentration of F343 reached 119 g/L after fermentation for 30 h. The genetic diversity between F343 and its ancestors was also evaluated by amplified fragment length polymorphism analysis. Results suggest that the phenotypes for both thermotolerance and l-glutamic acid production in F343 were evolved.





Similar content being viewed by others
References
Bona R, Moser A (1997) Modelling of the l-glutamic acid production with Corynebacterium glutamicum under biotin limitation. Bioprocess Biosyst Eng 17:139–142. doi:10.1007/PL00008956
Cao X, Song Q, Wang C, Hou L (2010) Genome shuffling of Hansenula anomala to improve flavour formation of soy sauce. World J Microbiol Biotechnol. doi:10.1007/s11274-010-0477-5
Dai MH, Shelley DC (2004) Genome shuffling improves degradation of the anthropogenic pesticide pentachlorophenol by Sphingobium chlorophenolicum ATCC 39723. Appl Environ Microbiol 70:2391–2397. doi:10.1128/AEM.70.4.2391-2397.2004
Del Cardayré SB, Tobin MS, Willem PC et al (2000) Evolution of whole cells and organisms by recursive sequence recombination. International patent application no. WO 00/04190
Delaunary S, Lapujade P, Engasser JM, Goergen JL (2002) Flexibility of the metabolism of Corynebacterium glutamicum 2262, a glutamic acid-producing bacterium, in response to temperature upshocks. J Ind Microbiol Biotechnol 28:333–337
Delaunay S, Gourdon P, Lapujade P et al (1999) An improved temperature-triggered process for glutamate production with Corynebacterium glutamicum. Enzym Microb Technol 25:762–768. doi:10.1016/S0141-0229(99)00120-9
Fudou R, Jojima Y, Seto A, Yamada K, Kimura E, Nakamatsu T, Hiraishi A, Yamanaka S (2002) Corynebacterium efficiens sp. nov., a l-glutamic acid-producing species from soil and vegetables. Int J Syst Evol Microbiol 52:1127–1131
Gong J, Zheng H, Wu Z, Chen T, Zhao X (2009) Genome shuffling: progress and applications for phenotype improvement. Biotechnol Adv 27:996–1005. doi:10.1016/j.biotechadv.2009.05.016
Hermann T (2003) Industrial production of amino acids by coryneform bacteria. J Biotechnol 104:155–172. doi:10.1016/s0168(03)00149-4
Hida H, Yamada T, Yamada Y (2006) Genome shuffling of Streptomyces sp. U121 for improved production of hydroxycitric acid. Appl Microbiol Biotechnol 73:1387–1393. doi:10.1007/s00253-006-0613-1
Hoischen C, Krämer R (1988) Evidence for an efflux carrier system involved in the secretion of glutamate by Corynebacterium glutamicum. Arch Microbiol 151:342–347. doi:10.1007/BF00406562
Hou LH (2009) Novel methods of genome shuffling in Saccharomyces cerevisiae. Biotechnol Lett 31:671–677. doi:10.1007/s10529-009-9916-5
Ikeda M, Nakagawa S (2003) The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl Microbiol Biotechnol 62:99–109. doi:10.1007/s00253-003-1328-1
Jin QC, Jin ZH, Xu B, Wang Q, Lei Y, Yao S, Cen P (2008) Genomic variability among high pristinamycin-producing recombinants of Streptomyces pristinaespiralis revealed by amplified fragment length polymorphism. Biotechnol Lett 30:1423–1429. doi:10.1007/s10529-008-9701-x
Kalinowski J, Bathe B, Bartels D et al (2003) The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate derived amino acids and vitamins. J Biotechnol 104:5–25. doi:10.1016/S0168-1656(03)00154-8
Kaneko H, Sakaguchi K (1979) Fusion of protoplasts and genetic recombination of Brevibacterium flavum. Agric Biol Chem 43:1007–1013
Katsumata R, Ozaki A, Oka T, Furuya A (1984) Protoplast transformation of glutamate-producing bacteria with plasmid DNA. J Bacteriol 159:306–311
Kimura E (2005) l-Glutamate production. In: Eggeling L, Bott M (eds) Handbook of Corynebacterium glutamicum. CRC press, Florida, pp 439–457
Kimura E, Kikuchi Y, Kawahara Y, Goto S, Kurahashi O, Nakamatsu T (2002) Stress-resistant microorganism and method of producing fermentative product. US patent 6,338,956
Marquet M, Uribelarrea JL, Huchenq A, Lane-elle G, Goma G (1986) Glutamate excretion by Corynebacterium glutamicum: a study of glutamate accumulation during a fermentation course. Appl Microbiol Biotechnol 25:220–223. doi:10.1007/BF00253652
Momose H, Takagi T (1978) Glutamic acid production in biotin rich media by temperature sensitive mutants of Brevibacterium lactofermentum, a novel fermentation process. Agric Biol Chem 42:1911–1917
Muffler A, Betterman S, Haushalter M, Horlein A, Neveling U, Schramm M, Sorgenfrei O (2002) Genome-wide transcription profiling of Corynebacterium glutamicum after heat shock and during growth on acetate and glucose. J Biotechnol 98:255–268
Nešvera J, Pátek M (2011) Tools for genetic manipulations in Corynebacterium glutamicum and their applications. Appl Microbiol Biotechnol 90:1641–1654. doi:10.1007/s00253-011-3272-9
Nishio Y, Nakamura Y, Kawarabayasi Y et al (2003) Comparative complete genome sequence analysis of the amino acid replacements responsible for the thermostability of Corynebacterium efficiens. Genome Res 13:1572–1579
Nunheimer TD, Birnbaum J, Ihnen ED, Demain AL (1970) Product inhibition of the fermentative formation of glutamic acid. Appl Microbiol 20:215–217
Patnaik R, Louie S, Gavrilovic V et al (2002) Genome shuffling of Lactobacillus for improved acid tolerance. Nat Biotechnol 20:707–712
Peberdy JF (1980) Protoplast fusion—a tool for genetic manipulation and breeding in industrial microorganisms. Enzym Microb Technol 2:23–29. doi:10.1016/0141-0229(80)90004-6
Santos CNS, Stephanopoulos G (2008) Combinatorial engineering of microbes for optimizing cellular phenotype. Curr Opin Chem Biol 12:168–176
Shiio I, Otsuka SI, Katsuya M (1962) Effect of biotin on the bacterial formation of glutamic acid. I Glutamate formation and cellular permeability of amino acids. J Biochem 51:56–62
Shiratsuchi M, Kuronuma H, Kawahara Y, Yoshihara Y, Miwa H, Nakamori S (1995) Simultaneous and high fermentative production of lysine and l-glutamic acid using a strain of Brevibacterium lactofermentum. Biosci Biotechnol Biochem 59:83–86
Sun Z, Yu Z, Yang Y, Jin H, Yang H (1989) Fermentative production of l-glutamic from rice hydrolysate by temperature-sensitive mutant Corynebacterium crenatum N1. Ind Microbiol 19:9–14
Takinami K, Yamada Y, Okada H (1965) Biochemical effects of fatty acids and its derivatives on l-glutamic acid fermentations. III Biotin-tween 60 relationship in the accumulation of l-glutamic acid and the growth of Brevibacterium lactofermentum. Agric Biol Chem 29:351–359
Takors R, Bathe B, Rieping M, Hans S, Kelle R, Huthmacher K (2007) Systems biology for industrial strains and fermentation processes—example: amino acids. J Biotechnol 129:181–190. doi:10.1016/j.jbiotec.2007.01.031
Vertès AA, Inui M, Yukawa H (2005) Manipulating Corynebacteria, from individual genes to chromosomes. Appl Environ Mirobiol 71:7633 –7642. doi:10.1128/AEM.71.12.7633-76422005
Vos P, Hogers R, Bleeker M et al (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wendisch VF, Bott M, Kalinowski J, Oldiges M, Wiechert W (2006) Emerging Corynebacterium glutamicum systems biology. J Biotechnol 124 :74–92. doi:10.1016/j.jbiotec.2005.12.002
Xu B, Jin Z, Wang H, Jin Q, Jin X, Cen P (2008) Evolution of Streptomyces pristinaespiralis for resistance and production of pristinamycin by genome shuffling. Appl Microbiol Biotechnol 80:261–267. doi:10.1007/s00253-008-1540-0
Zhang YX, Perry K, Vinci VA et al (2002) Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature 415:644–646
Zhang CY, Shi ZP, Gao P, Duan Z, Mao Z (2005) On-line prediction of products concentrations in glutamate fermentation using metabolic network model and linear programming. Biochem Eng J 25:99–108. doi:10.1016/j.bej.2005.03.012
Zhao M, Dai CC, Guan XY, Tao J (2009) Genome shuffling amplifies the carbon source spectrum and improves arachidonic acid production in Diasporangium sp. Enzym Microb Technol 45:419–425. doi:10.1016/j.enzmictec.2009.08.012
Acknowledgments
This work was supported by grants from the National High Technology Research and Development Program of China (No. 2006AA020301-09).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zheng, P., Liu, M., Liu, Xd. et al. Genome shuffling improves thermotolerance and glutamic acid production of Corynebacteria glutamicum . World J Microbiol Biotechnol 28, 1035–1043 (2012). https://doi.org/10.1007/s11274-011-0902-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-011-0902-4