Abstract
The effects of a maize knotted1 (kn1) gene under the control of the cauliflower mosaic virus 35S promoter on genetic transformation efficiencies of six citrus genotypes were tested. The kn1 gene construct was used to transform ‘Pineapple’, ‘Hamlin’, ‘Sucarri’ and ‘Valencia’ sweet orange [Citrus sinensis (L.) Osbeck], ‘Carrizo’ Citrange [Citrus sinensis (L.) Osbeck × Poncirus trifoliate (L.) Raf.] and Eureka lemon [Citrus. limon (L.) Burm.f.] via Agrobacterium tumefaciens-mediated infection. Our results demonstrate that expression of the kn1 gene enhances transformation efficiencies from 3 to 15 fold compared to a control vector, 3–11 fold relative to the highest transformation efficiencies reported for these citrus genotypes. Stable incorporations of T-DNA into the citrus genome have been confirmed with both histochemical staining of GUS activity and molecular analyses. The majority of kn1 over-expressing citrus plants grow and develop normally at young seedling stages, similar to those of the wild type plants. With all six genotypes of citrus tested including Eureka lemon, a cultivar difficult to be transformed, our results demonstrate that the kn1 gene may provide an effective molecular tool to enhance genetic transformation efficiencies of various citrus varieties. High transformation efficiency of citrus is of great importance for large scale characterization of gene functions and also cultivar development via transgenic and genome editing technologies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Citrus is commercially grown over a wide range of climatic conditions and soil types where crops are exposed to a variety of abiotic and biotic stresses that often negatively affect fruit quality and productivity. Important abiotic stresses include excess soil acidity or alkalinity, drought, and temperature extremes, while insects and diseases represent major biotic stresses (Pena et al. 2007). Some diseases, such as Hunglongbing (HLB) and citrus canker, have spread to citrus production areas throughout the world. For instance HLB has caused huge losses to the citrus industries in the US, China and many other countries (Bové 2006; Wang and Trivedi 2013). While no effective HLB control methods or HLB resistance citrus cultivars have been commercialized, transgenic plant technologies have been recognized as a powerful tool to control the HLB disease.
An efficient genetic transformation protocol is a key step for using transgenic technologies to improve citrus quality, productivity, and resistance to abiotic and biotic stresses such as HLB. Several citrus transformation protocols have been previously published. As Donmez et al. (2013) pointed out, however, that “transformation efficiency (for citrus) is generally low and protocols are only effective on certain species or even cultivars”. One major limitation associated with existing protocols when used on economically important citrus species is likely low plant regeneration frequencies. Another challenge to using transgenic plant technology in citrus is the long juvenile phase of immature tissues that ranges from 5 to 20 years, which greatly delays analysis of the effects of candidate genes. On the other hand, transgenic plants produced from mature tissue via genetic transformation can drastically shorten flowering and fruiting time and thus speed up field evaluation studies. Cervera et al. (1998) have shown that greenhouse-grown transgenic plants derived from mature tissues can fruit in 14 months. However, efforts have been made to genetically transform mature citrus tissues but the efficiencies are generally extremely low (Marutani-Hert et al. 2012).
The maize homeobox gene knotted1 (kn1) encodes an AP2/ERF transcription factor (Hake et al. 2004; Moon and Hake 2011). It has also been shown that the kn1 mRNA can be transported bidirectionally via phloem cells in plants (Duan et al. 2014). KN1 may regulate many processes including hormone concentrations and growth and development of floral, shoot meristem and leaf organs (Bolduc et al. 2012; Hake et al. 2004; Hay and Tsiantis 2010; Sano et al. 2005). Ipt, a cytokinin biosynthetic gene cloned from Agrobacterium, has also been shown to stimulate shoot regeneration (Li et al. 1992; Smigocki and Owens 1988, 1989; Strabala et al. 1989, 1996). Further studies have shown the ipt gene may be used as a positive selectable marker for transformation in citrus, tobacco, rice, sweet pepper, aspen and apricot (Ballester et al. 2007, 2008; Ebinuma and Komamine 2001; Ebinuma et al. 1997; Endo et al. 2001, 2002; Kunkel et al. 1999; López-Noguera et al. 2009; Mihálka et al. 2003; Peng et al. 2015). Similar to the ipt gene, we have previously reported that the kn1 gene can be used as a positive selection marker for transgenic tobacco plants (Luo et al. 2006), The utility of the kn1 gene in genetic transformation of higher plants has also been shown in Phalaenopsis amabilis and Jatropha curcas (Pei et al. 2010; Semiarti et al. 2007).
Previously, using a sonication-assisted Agrobacterium-mediated transformation method combined with vacuum infiltration, Oliveira et al. (2009) reported an 8.4 % transformation efficiencies for juvenile tissues of ‘Pineapple’ orange. Dutt and Grosser (2009) described an improved protocol for genetic transformation of juvenile explants of ‘Carrizo’ citrange and ‘Hamlin’ orange. The authors optimized several parameters including pre-incubation treatment, OD values of Agrobacterium cells and Agrobacterium-explant co-cultivation durations. They reported optimization conditions for both cultivars and achieved the maximum transgenic plant production, 47 % transformation efficiency for ‘Carrizo’ and 25 % for ‘Hamlin’. However, this type of methods is likely species or cultivar dependent.
The aim of this study was to determine the effects of the kn1 gene on genetic transformation efficiencies of juvenile citrus explant tissues using six citrus cultivars, ‘Carrizo’ citrange [Citrus sinensis (L.) Osbeck × Poncirus trifoliata (L.) Raf.], ‘Pineapple’, ‘Hamlin’, ‘Succari’, ‘Valencia’ sweet orange [Citrus sinensis (L). Osbeck] and ‘Eureka’ lemon [Citrus. limon (L.) Burm.f.]. Here, we report that transformation efficiencies of the six citrus cultivars can be enhanced drastically from 3 to 15 fold if the kn1 gene is used. Also, no significant changes in morphology or growth habits have been observed in the kn1 transgenic citrus plants at seedling stages, which is different from kn1 over-expressing transgenic plants previously described in the literature. With all citrus cultivars we have tested including a lemon cultivar, we conclude that the kn1 gene can be useful to enhance genetic transformation efficiencies of various citrus varieties.
Materials and methods
Plant Materials
‘Carrizo’ Citrange [Citrus. sinensis (L.) Osbeck × Poncirus trifoliata (L.) Raf.] seeds were purchased from TreeSource CitrusNursery (504 N Kaweah Ave, Exeter, CA 93221 U.S.), seeds for ‘Pineapple’, ‘Succari’, and ‘Hamlin’ sweet orange [Citrus sinensis (L.) Osbeck] were provided by the University of Florida and Valencia orange [Citrus sinensis (L.) Osbeck] and Eureka lemon [Citrus. limon (L.) Burm.f.] seeds were purchased from Pearson Ranch California Oranges (1018 W. Teapot Dome Ave. Porterville, CA 93257). External seed coats were removed and seeds were treated with 75 % alcohol for 40–60 s and 1 % sodium hypochlorite for 20 min, and rinsed four times with sterile distilled water. Internal seed coats were then removed under sterile conditions and seeds were cultured in vitro on MS media (Murashige and Skoog 1962) with 30 g/l sucrose, 7 g/l of agar and pH 5.7. Thirty-day-old seedlings were used as a source of explant tissue for transformation. The internodal stem segments about 1 cm in length were harvested from these seedlings and used for Agrobacterium infection as reported previously (Dutt and Grosser 2009).
Vector and Agrobacterium for transformation
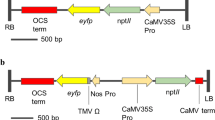
Agrobacterium tumefaciens EHA 105 carrying the binary vector pBin19 with the 35S Promoter::kn1 (Luo et al. 2006) or the SAUR Promoter::ipt (Li et al. 1991, 1992) and 35S-nptII::uidA gene plus the vector with only the 35S-nptII::uidA gene as control vector (Zheng et al. 2007). In these vectors, the nptII gene (the kanamycin resistance gene) served as a marker gene for the selection of transgenic plants during transformation. The uidA (β-glucuronidase gene) was used as a report gene for the screening of positive transgenic plants.
Agrobacterium tumefaciens EHA 105 strains hosting various pBin 19 vectors were cultured for 40–42 h at 28 °C on LB solid medium containing 100 mg/l kanamycin and 50 mg/l rifampicin. Single colonies were transferred to 2 ml LB liquid medium with 100 mg/l kanamycin and cultivated under 180 rpm, 28 °C for 24 h. After that, 1–2 ml(s) cultivated bacteria was added to 40 ml LB liquid medium with 100 mg/l kanamycin and cultivated to an OD of around 0.6 and then centrifuged at 5000 rpm at room temperatures for 15 min. Prior to plant infection, the bacterial culture was re-suspended in liquid co-cultivation medium (CCM) consisting of MS salts, 0.1 mg/l thiamine hydrochloride, 0.5 mg/l pyridoxine hydrocholoride, 0.5 mg/l nicotinic acid, 2 mg/l glycine, 100 mg/l inositol, 20 mg/l acetosyringone (AS), 30 g/l of sucrose, and 3 mg/l benzylaminopurine (BAP) for ‘Carrizo’, ‘Succari’, ‘Eureka’, and ‘Valencia’, 3 mg/l BAP, 0.1 mg/l 1-naphthlcetic acid (NAA) and 1 mg/l 2,4-dichlorophenoxyacetic acid (2,4-D) for ‘Hamlin’ and ‘Pineapple’.
Infection and shoot regeneration
Internodal stem segments (1 cm in length) from 30 day old citrus seedlings were incubated in the Agrobacterium cell suspension basically as described by Yang et al. (2011). After being blotted dry on sterilized filter paper, explants were placed horizontally in Petri dishes containing solid CCM and incubated in the dark. Three days after, the internodal stem segments were blotted dry on sterile filter paper and transferred onto shoot regeneration medium (SRM) containing MS salts, 0.1 mg/l thiamine hydrochloride, 0.5 mg/l pyridoxine hydrochloride, 0.5 mg/l nicotinic acid, 2 mg/l glycine, 100 mg/l inositol, 30 g/l of sucrose, 7 g/l of agar. For ‘Carrizo’, 3 mg/l BAP was added, but 1 mg/l BAP was used for the rest of other four sweet orange cultivars. In the case of ‘Eureka’ lemon, we used 0.5 mg/l BAP. All SRM were supplemented with 75 mg/l of kanamycin and 150 mg/l of timentin. The explant tissues were transferred onto fresh SRM plates every 3 weeks, and cultured under full spectrum light (45 μEm−2 s−1) for 16 h per day at 26 ± 2 °C. GUS positive shoots regenerated from the kanamycin containing medium were grafted on 14 days old ‘Carrizo’ citrange rootstocks. One to two months later, the established shoots were re-grafted on potted sour orange under greenhouse conditions. We also rooted GUS positive ‘Carrizo’ shoots in root-inducing medium, which contained half-strength MS salts, 0.1 mg/l thiamine hydrochloride, 0.5 mg/l pyridoxine hydrochloride, 0.5 mg/l nicotinic acid, 2 mg/l glycine, 100 mg/l inositol, 0.5 mg/l NAA, 0.1 mg/l IBA and 0.5 g/l activated charcoal.
GUS histochemical assays and data analysis
Histochemical assays of GUS activity were carried out in a solution consisting of 100 mM potassium phosphate buffer, pH 7.0, 10 mM Na2EDTA, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 0.1 % triton X-100, 1 g/l X-gluc (5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid) at 37 °C for 16 h. After 45 days cultured on the shoots induction medium, small leaf slices or entire buds of kanamycin resistant shoots that were longer than 3 mm were used for histochemical GUS staining. The plant tissues were distained in ethanol to remove chlorophylls and other pigments prior to being visually inspected and photographed. Transformation efficiency is defined as the average number of GUS positive and PCR verified shoots per explant inoculated × 100 %.
All data were analyzed with the SPSS statistical software and Duncan’s multiple range test at P ≤ 0.05.
Molecular confirmation of transgenic plants
Genomic DNA was extracted from leaves of GUS positive and wild type citrus plants. To avoid contaminations of Ti-plasmid DNA from Agrobacterium remained in citrus plant tissues, the genomic DNA isolated was fractioned on 0.8 % (w/v) agarose gel with the pBin19-35S::kn1 Ti-plasmid DNA loaded on the side as a reference. Large-sized-genomic DNAs (about 20–25 kb that is much larger than the Ti-plasmid DNA) were recovered from the agarose gels and used as templates of PCR reactions (Chen et al. 2006). These DNA templates were analyzed by PCR for the uidA gene within the T-DNA region and the tetracycline resistance (tetR) gene that is outside the T-DNA region of the Ti-plasmid. PCR reaction solution was 20 μl containing 1× PCR buffer (Takara, Japan), 1.5 mM MgCl2, 0.2 mM dNTPs, 0.2 μl e2TAK DNA polymerase (Takara, Japan), 0.25 μM of each primer and 500 ng DNA. The amplification condition started with an initial denaturation step at 98 °C for 5 min, followed by 35 cycles of 98 °C for 10 s, special annealing temperature for 5 s, and 72 °C extension plus a final extension at 72 °C for 10 min. The primers used to amplify the uidA gene are gus-F and gus-R, The predicted size of the uidA DNA segment amplified is 823 bp. the primers used to amplify the tetracycline gene tetR gene are Tet-F and Tet-R, the predicted size of the amplified tetR DNA segment is 552 bp. PCR products were separated by electrophoresis on 1 % (w/v) agarose gels. The PCR primers used: gus-F: 5′-ATACCGAAAGGTTGGGC AGG; gus-R 5′-TCACCACGATGCCATGTTCA; tet-F: 5′-GACGAC TGGCGCTCATTTCT; tet-R: 5′-GCATGAAAAAGCCCGTAGCG.
Expression analysis of kn1 genes in transgenic plant leaves
Total RNA was isolated from leaves of transgenic and wild type plants using RNeasy Plant Mini Kit (QIAGEN) according to the manual. cDNA synthesis was done with iScript™ cDNA Synthesis Kit (Bio-Rad).
Real time quantitative PCR was performed to analyze the expression levels of the transgenes using SsoFast™ qPCR Supermixes with EvaGreen (Bio-Rad). The reaction system included 20 μl of 1× Supermixes, 0.5 μM each primer for kn1, 200 ng cDNA. The qPCR primers used are: qKN1-F: 5′-GAAGCACCATCTCCTGAA; qKN1-R: 5′-CCACCTTCTGAG TCTCTG. Reactions were performed in 40 cycles of 30 s at 95 °C, 5 s at 95 °C, 20 s at 59 °C and 31 s at 72 °C. We used expression of the ActB gene as a reference gene for the qPCR assays because Yan et al. (2012) have recommended it as a suitable reference gene to normalize mRNA levels in qPCR analysis of diverse cultivars and tissues based on extensive studies. The qPCR premiers for the ActB gene: ACTB-F: 5′-TCTCTTGAACCTGTCCTTGGA; ACTB-R: 5′-AGTGCC GATACGCTGTCTA. Each sample had three replicates. Relative expression levels were shown as the ratio of the transgenic kn1 gene to the endogenous ActB gene (Yan et al. 2012).
Results and discussion
Kn1 overexpression drastically increases transformation efficiencies of six citrus cultivars tested
KN1 is a transcription factor protein involved in the establishment and maintenance of plant meristems (Bolduc and Hake 2009). The kn1 gene has been previously used to improve shoot regeneration and transformation in tobacco (Luo et al. 2006; Tamaoki et al. 1997), jatropha (Pei et al. 2010), orchid (Semiarti et al. 2007) and barley (Williams-Carrier et al. 1997). To test the effect of the kn1 overexpression on genetic transformation of citrus plants, we used A. tumefaciens stains hosting a control vector (uidA and kanamycin resistance genes), the kn1 gene plus the uidA and kanamycin resistant genes (nptII) to transform six cultivars of citrus, ‘Carrizo’ citrange, ‘Pineapple’, ‘Hamlin’, ‘Succari’, ‘Valencia’ sweet orange and ‘Eureka’ lemon. We also used an ipt gene plus the uidA and nptII genes to transform ‘Carrizo’ citrange, ‘Pineapple’, ‘Hamlin’ and ‘Succari’ orange. We did three to five transformation experiments using hundreds of epicotyl explants for each construct and each citrus cultivar. As shown in Fig. 1a, b, the use of the kn1 or ipt genes drastically increased transformation/shoot regeneration efficiencies from epicotyl segments of the 30 days old seedlings of these citrus cultivars.
Effects of kn1 and ipt overexpression on efficiencies of citrus transformation. a, b Shoots regenerated from the explants infected with Agrobacterium containing pBin19-35S::kn1, pBin19-SAUR::ipt and control vector (a ‘Pineapple’, b ‘Hamlin’), showing that kn1 and ipt gene enhance citrus transformation efficiencies. c Transgenic SAUR::ipt ‘Pineapple’ shoots with increase in bud/shoot regeneration efficiency. d Histochemical staining of GUS activity in kn1 transgenic shoots and leaves. e Grafted Pineapple wild-type (the one on the left) and transgenic 35S::kn1 ‘Pineapple’ plants (the two plants on the right), these plants were grafted at the same time (1 month old). f A leaf from wild-type Pineapple citrus. g A leaf from 35S::kn1 Pineapple H10, with no differences observed when compared to wild-type ‘Pineapple’ leaves. h A leaf from 35S::kn1 Pineapple H7 plant, with non-smooth surfaces when compared to the leaves from wild type Pineapple citrus plants. i A leaf from kn1 transgenic Pineapple H2 with no petiole wings. j Three month old wild type ‘Carrizo’ citrus plant (left) and kn1 transgenic ‘Carrizo’ citrus plants (middle and right), showing the kn1 transgenic ‘Carrizo’ citrus plants root and grow normally. k Eight month old ‘Succari’ wild type (left) and kn1 transgenic plants (right) that were grafted on sour orange rootstocks, showing no significant difference between wild-type and kn1 transgenic plants
Table 1 summarizes the effects of the kn1 and ipt genes and control vector on genetic transformation efficiencies of the six citrus cultivars tested. Transformation efficiencies were calculated by dividing the number of independent GUS positive, PCR confirmed shoots with the total number of explants inoculated and then multiplied by 100 %. Compared to the control vector with a uidA gene, we found that the use of the ipt gene increased transformation efficiency by 4.8 fold for ‘Pineapple’ orange, 5.0 fold for ‘Hamlin’ orange, 4.3 fold for ‘Succari’ orange and 8.6 fold for ‘Carrizo’ citrange. Using the kn1 gene, we observed increases in transformation efficiency for 4.9 fold for ‘Pineapple’ orange, 5.1 fold for ‘Hamlin’ orange, 6.4 fold for ‘Succari’ orange, 15.0 fold for ‘Carrizo’ citrange, 4.2 fold for ‘Valencia’ orange and 3.3 fold for ‘Eureka’ lemon. Statistical analyses confirm that the differences in transformation efficiency between the kn1 or ipt vector and the control vector are significant.
In Table 1, we also listed the highest genetic transformation efficiency reported previously in the literature for a comparison. For ‘Pineapple’ sweet orange, the transformation efficiency for the control vector (pBin19 35S::nptII::uidA gene) was 19.2 % in our hand and the highest transformation efficiency reported in the literature was 8.4 % (Oliveira et al. 2009). When the kn1 gene was used, the efficiency was 95 %. In the case of ‘Hamlin’ sweet orange and ‘Carrizo’ citrange, we obtained 13.5 and 10.3 % transformation efficiencies, respectively, when the control vector was used. The highest transformation efficiencies reported for these two cultivars were 25 and 47 % (Dutt and Grosser 2009). Using the kn1 gene, we observed 69 and 155 % transformation efficiencies for these two cultivars, respectively. For ‘Valencia’ orange, a commercially important cultivar, we observed 14 % transformation efficiency using the control vector and the previously reported transformation efficiencies were 2 % (Dutt et al. 2009) to 24 % (Boscariol et al. 2003; Khan et al. 2012). With the kn1 gene, we observed 61 % transformation efficiency.
The differences in the transformation efficiencies between ours without the use of the kn1 or ipt genes and those reported by others previously (see Table 1) could be due to differences in vigor for explants and Agrobacterium cells or other variations. However, compared with the highest transformation efficiencies reported in the literature and also the control vectors under identical conditions in our own study, the use of the kn1 gene always drastically enhanced the transformation efficiency of the citrus cultivars tested (Table 1).
Although reasonably high transformation efficiencies had already been achieved previously for some citrus cultivars (see Table 1), genetic transformation for many other citrus species or cultivars including commercially used Satsuma Mandarin (Citrus unshiu Marc.), lemon [Citrus limon (L.) Burm. f.], Ponkan (Citrus reticulata Blanco) has been difficult (Dutt and Grosser 2010; Dutt et al. 2009; Khawale et al. 2006). For instance, transformation efficiencies of lemon cultivars are normally 1 % to 3 % (Dutt et al. 2009; Ghorbel et al. 2001). With the use of the kn1 gene, we have showed that the transformation efficiency for Eureka Lemon can be as high as 28 % (Table 1). More recently, Xiao et al. (2014) reported an efficient Agrobacterium rhizogenes-mediated transformation method for Poncirus trifoliate, with a transformation efficiency of 34 %. Their procedure requires 25 weeks to produce transgenic shoots while our method needs 12 weeks.
Taken all together, our results demonstrate that kn1 gene is very effective in enhancing genetic transformation efficiencies of all six citrus cultivars tested, with increases in transformation efficiency from 3 to 15 fold from the control vector without the use of the transgenic kn1 gene, or 3–11 fold increase when compared to the highest transformation efficiencies previously published. Further, these six citrus cultivars tested also include a lemon cultivar. Lemon has been generally regarded difficult to be transformed with Agrobacterium (Dutt et al. 2009; Ghorbel et al. 2001).
Verification of the stable incorporation of the transgenes into citrus genomes
Because all Ti-plasmid vectors we used in this study contain the 35S::nptII::uidA fusion gene, in addition to the kanamycin resistance, we used histochemical staining of the GUS activity to confirm stable incorporations of the transgenes into the citrus genome in independently produced shoots of citrus (Fig. 1d). We also used a PCR technique to further verify the stable integration of transgenes into the citrus genome of kanamycin resistant, GUS positive shoots. Since the genomic DNA from T0 generation transgenic citrus plant tissues were used for PCR reactions, it is possible to observe false-positive PCR results because of the Ti-plasmid DNA from residual Agrobacterium cells in the plant tissues. Using the method we described previously (Chen et al. 2006), we eliminated possible contaminations of Ti-plasmid DNA from residual Agrobacterium cells by fractionating the citrus genomic DNA isolated from T0 transgenic plant tissues on agarose gels with pBin19-35S::kn1 plasmid DNA as a reference. As shown in Lane 2–9 of Fig. 2a, we were able to separate high molecular weight citrus genomic DNA from the Ti-plasmid DNA on agarose gels. We then recovered high molecular weight citrus genomic DNA with no plasmid T-DNA from the agarose gel and used them as PCR templates. We isolated and purified genomic DNA from randomly selected 12 GUS-positive lines of citrus shoots derived from epicotyl segments infected with Agrobacterium strain containing the kn1 gene. We demonstrated that all 12 GUS-positive lines were PCR positive for the kn1 gene. Four representative PCR products from these 12 lines were shown in Fig. 2b, demonstrating the presence of the uidA gene (Lanes 4, 6, 8 and 10).
PCR Confirmation of stable incorporation of transgenes into the citrus genome. a Genome DNA isolated from GUS positive citrus tissues were fractioned on an agarose gel. DNA in the marked regions were recovered for PCR reactions. Lane M Molecular weight mark of lambda DNA digested with EcoRI and HindIII, Lane 1 pBin19-35S::kn1 plasmid DNA, Lanes 2–9 Undigested genomic DNA extracted from transgenic plants. b PCR reactions were performed as described in “Materials and methods” with primer sequences for the uidA gene (within the T-DNA region and should be incorporated into the plant genome) and for the tetR gene (outside the T-DNA region, should not be incorporated into the plant genome). Lane M: Molecular weight marker. The lane bands 1, 3, 4, 6, 8 and 10 are for the uidA gene, lane bands 2, 5, 7, 9 and 11 are for the tetR gene, Lanes 1 and 2 Positive control (pBin19-35S::kn1 plasmid DNA as template) for the uidA and tetR gene, respectively. Lane 3: Negative control (Wild-type Pineapple citrus plant DNA as template) for the uidA gene, Lane 4–10: GUS positive ‘Pineapple’ citrus line 2, 3, 7 and 10 with the presence of the GUS gene and the absence of the tetR gene, demonstrating that these plant lines are transgenic
To further verify that the presence of transgenes in these GUS positive citrus shoot lines (for examples see Fig. 1d) were not from Ti-plasmid DNA in residual Agrobacterium cells in T0 plant tissues, we performed an additional PCR analysis using primers for the tetracycline resistance (tetR) gene located in the backbone region of the Ti-plasmid DNA. Our results show that no tetR was detected from the same genomic DNA templates for which the uidA gene was identified (Fig. 2b shows four representative PCR reactions). These results demonstrate that our technique (Chen et al. 2006) is effective to eliminate the Ti-plasmid DNA from the template DNAs isolated GUS positive citrus tissues, and therefore the uidA positive signals observed in the representative plant lines were due to stable incorporations of the transgenes in the citrus genome. Further, some transgenic plants were further confirmed with qPCR for the expression of the kn1 gene.
Expression of the Kn1 genes and their effects on citrus plant growth and development
When using the ipt gene for citrus transformation, we observed that many of the ipt transgenic citrus shoots displayed a bushy phenotype with small shoots or buds (Fig. 1c) and poor root systems (data not shown). We therefore did not characterize the ipt citrus plants further. On the other hand, the kn1 transgenic citrus shoots were normal in growth and development patterns at young seedling stages when compared to the wild type plants. We randomly selected 47 kn1 transgenic shoots for further analysis. We grafted 42 kn1 shoots onto rootstocks of sour orange (Citrus aurantium L.). Figure 1e shows a wild type shoot grafted onto a sour orange rootstock (left), two kn1 ‘Pineapple’ sweet orange shoot grafted onto a rootstock (middle and right). All of these grafted shoots grew normally except two plant lines showing minor alterations in leaf morphology. Kn1-Pineapple H7 produced non-smooth leaf surfaces (Fig. 1h) and Kn1-Pineapple H2 failed to develop petiole wings (Fig. 1i). Other than these two minor changes, we did not notice any other differences in the kn1-Pineapple H2 and kn1-Pineapple H7 trees when compared to the rest of the kn1 transgenic lines or wild type plants. We also rooted five representative kn1 transgenic shoots of ‘Carrizo’ citrange and they produced normal roots, indistinguishable from the wild type plants when rooted under the same conditions (Fig. 1j).
Along with the 42 grafted lines of kn1 transgenic shoots on sour orange rootstocks, we also grafted the wild type shoots as controls. We did not notice any difference in grafting behavior between the kn1 transgenic and wild type shoots. Both rooted and grafted kn1 transgenic plants grew and developed normally when compared to the wild types plants. Except for the non-smooth leaf surfaces observed in kn1-Pineapple H7 and missing petiole wings on the kn1-Pineapple H2 plants, the 47 kn1 transgenic citrus plant lines developed and grew normally (Fig. 1j, k). We have not observed any other drastically altered growth and development patterns in all 47 transgenic kn1 citrus trees that are now 9–12 month old.
We also determined expression levels of the kn1 gene in 6 representative transgenic citrus plant liness. Table 2 shows that the expression level of the kn1 gene in kn1 Pineapple H7 transgenic trees was about 17.5 % of that of the internal reference gene, ActB (an actin gene in citrus). Yan et al. (2012) examined seven candidate reference genes (18SrRNA, ActB, rpII, UBQI, UBQ10, GAPDH and TUB) and concluded that ActB is one of the three best reference genes for normalizing mRNA levels in qPCR analysis of diverse cultivars and tissues of citrus plants. Our qPCR results show that the expression level of the kn1 gene in Pineapple H2 line was 2.7 % of that of the ActB gene. The rest of the four transgenic plant lines had 0.1 % or less than 0.1 % of the ActB expression levels (Table 2). Kn1 Pineapple H7 transgenic tree produced non-smooth surface leaves while Pineapple H2 transgenic tree is petiole wing-less (Fig. 1f–i). The rest of the transgenic tree lines were morphologically indistinguishable from wild type plants. The fact that most kn1 transgenic plant lines had no changes in growth and developmental patterns also support that kn1 mediated enhancement of citrus transformation efficiency may not require high expression levels of the kn1 gene.
It has been well documented that overexpression of the kn1 gene causes drastic morphological alterations in transgenic plants including loss of apical dominance and increases in adventitious shoot formation, reduction in shoot elongation, and abnormal leaf and floral morphologies, with many of these phenotypes being similar to those observed in ipt over-expressing plants (Sinha et al. 1993; Chuck et al. 1996; Tamaoki et al. 1997). When used as a positive selection marker or for enhancing transformation efficiencies in tobacco (Luo et al. 2006; Tamaoki et al. 1997), jatropha (Pei et al. 2010), orchid (Semiarti et al. 2007) or barley (Williams-Carrier et al. 1997), the transgenic kn1 gene overexpression also caused bushy phenotype, reduced shoot growth, or altered floral and leaf morphology. It is therefore somewhat surprising that overexpression of the kn1 gene can drastically enhance the transformation efficiencies of all six citrus cultivars tested but does not cause significant alterations in growth and developmental patterns at the early stage of citrus plant growth and development.
The 35S CaMV gene promoter is a strong and constitutive promoter when used in higher plants (Zheng et al. 2007). However, if the 35S promoter is used to drive an ipt gene (a cytokinin biosynthetic gene) expression in tobacco or other plant species, transgenic plants produced have low levels of the ipt expression (Smigocki and Owens 1988; Li unpublished data). This is because transgenic cells with high ipt expression produced high levels of cytokinins which inhibit shoot elongation. In this study, similar to the effects of the 35S::ipt gene, high expression levels of the 35S::kn1 gene in citrus cells may also prevent shoot elongation, which may explain why most transgenic citrus plants we produced have relatively low kn1 expression levels. On the other hand, small increases in transgenic KN1 protein in citrus cells can enhance shoot regeneration upon Agrobacterium-mediated transformation but low amounts of the KN1 protein in roots, stems and leaves may not be sufficient to cause morphological changes in vegetative organs of young citrus seedlings.
Although we have not observed drastic changes in growth and development patterns of the 1 year old kn1 transgenic citrus seedlings, we did observe some changes in leaf morphology because of the expression of the kn1 transgene, which is consistent with phenotypes observed in some kn1 mutant or transgenic plants (Moon and Hake 2011). Further, it is not known whether these kn1 citrus plants will display any alterations in flower and fruit growth and development at the reproductive stage.
To use the kn1 gene to improve genetic transformation efficiencies of citrus cultivars of interest, one needs to insert the kn1 gene into the Ti-plasmid vector containing a target gene(s) for citrus transformation. Once transgenic buds/shoots are obtained, the 35S::kn1 gene can be deleted using the gene deletor technology (Luo et al. 2007), or inactivated using CRISPR technologies (Sander and Joung 2014; Xiong et al. 2015). On the other hand, in many cases, removal or inactivation of the kn1 gene may not be necessary since we have not observed detrimental alterations in plant growth and development in 1 year old kn1 transgenic seedlings.
In summary, we have demonstrated that the use of the 35S::kn1 gene drastically enhances genetic transformation of juvenile tissues of six citrus cultivars tested, including a lemon cultivar, ranging from 3 to 11 fold increases in efficiency. We have also observed that the kn1 overexpression in citrus does not lead to significant alterations in leaf morphology, shoot elongation, root development and other developmental and growth processes at early stages of seedling growth. These results demonstrate that kn1 gene may be an effective tool to enhance genetic transformation of citrus cultivars or species that are difficult to transform as we have demonstrated in the case of lemon. We are currently conducting experiments to determine the effects of the kn1 overexpression in genetic transformation of mature citrus tissues. Our preliminary results suggest the kn1 gene can also be effective. If that can be achieved, the kn1 gene can be used to accelerate candidate gene evaluation or new cultivar development for citrus Huanglongbing and canker disease resistance and also for the enhancement of fruit yield and quality. High transformation efficiency of citrus plants, particularly mature citrus tissues, is crucial for large scale analyses of candidate gene functions and also cultivar development via transgene- and genome editing technologies (Ding et al. 2014; Dutt et al. 2014; Li 2013; van Nocker and Gardiner 2014; Xiong et al. 2015).
Abbreviations
- CaMV 35S:
-
Cauliflower mosaic virus 35S
- kn1 :
-
Knotted1 gene
- ipt :
-
Isopentenyl transferase
- BAP:
-
Benzylaminopurine
- NAA:
-
Naphthaleneacetic acid
- IBA:
-
Indole-3-butyric acid
- nptII :
-
Neomycin phosphotransferase
- MS medium:
-
Musashige and Skoog medium
- GUS:
-
β-Glucuronidase
- uidA :
-
β-Glucuronidase gene
- OD:
-
Optical density
- SAUR:
-
Small auxin-up RNA
- AS:
-
Acetosyringone
- MES:
-
2-(N-morpholino) ethanesulfonic acid
- X-gluc:
-
5-Bromo-4-chloro-3-indolyl-β-d-glucuronic acid
- CCM:
-
Co-cultivation medium
- SRM:
-
Shoot regeneration medium
References
Ballester A, Cervera M, Pena L (2007) Efficient production of transgenic citrus plants using isopentenyl transferase positive selection and removal of the marker gene by site-specific recombination. Plant Cell Rep 26:39–45
Ballester A, Cervera M, Pena L (2008) Evaluation of selection strategies alternative to nptII in genetic transformation of citrus. Plant Cell Rep 27:1005–1015
Bolduc N, Hake S (2009) The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 21:1647–1658
Bolduc N, Yilmaz A, Mejia-Guerra MK, Morohashi K, O’Connor D, Grotewold E, Hake S (2012) Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev 26:1685–1690
Boscariol RL, Almeida WAB, Derbyshire M, Mourao FAA, Mendes BMJ (2003) The use of the PMI/mannose selection system to recover transgenic sweet orange plants (Citrus sinensis L. Osbeck). Plant Cell Rep 22:122–128
Bové JM (2006) Huanglongbing: a destructive, newly-emerging, century-old disease of citrus. J Plant Pathol 88:7–37
Cervera M, Juarez J, Navarro A, Pina JA, Duran-Vila N, Navarro L, Pena L (1998) Genetic transformation and regeneration of mature tissues of woody fruit plants bypassing the juvenile stage. Transgenic res 7:51–59
Chen Y, Lu L, Deng W, Yang X, McAvoy R, Zhao D, Pei Y, Luo K, Duan H, Smith W, Thammina C, Zheng X, Ellis D, Li Y (2006) In vitro regeneration and Agrobacterium-mediated genetic transformation of euonymus alatus. Plant Cell Rep 25:1043–1051
Chuck G, Lincoln C, Hake S (1996) KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8:1277–1289
Ding J, Duan H, Zhao D, Deng Z, Yi G, Li Y (2014) Molecular strategies for addressing gene flow concerns over environmental stress tolerant transgenic plants. Crit Rev Plant Sci 33(2–3):265–277
Donmez D, Simsek O, Izgu T, Kacar YA, Mendi YY (2013) Genetic transformation in citrus. Sci World J 2013:491207
Duan X, Zhang W, Huang J, Zhao L, Ma C, Hao L, Yuan H, Harada T, Li T (2014) KNOTTED1 mRNA undergoes long-distance transport and interacts with movement protein binding protein 2C in pear (Pyrus betulaefolia). Plant Cell, Tissue Organ Cult 121:109–119
Dutt M, Grosser JW (2009) Evaluation of parameters affecting Agrobacterium-mediated transformation of citrus. Plant Cell, Tissue Organ Cult 98:331–340
Dutt M, Grosser JW (2010) An embryogenic suspension cell culture system for Agrobacterium-mediated transformation of citrus. Plant Cell Rep 29:1251–1260
Dutt M, Orbovic V, Grosser JW (2009) Cultivar dependent gene transfer into citrus using Agrobacterium. Proc Fla State Hortic Soc 122:85–89
Dutt M, Dhekney SA, Soriano L, Kandel R, Grosser JW (2014) Temporal and spatial control of gene expression in horticultural crops. Hortic Res 1:47
Ebinuma H, Komamine A (2001) MAT (Multi-Auto-Transformation) vector system. The oncogenes of Agrobacterium as positive markers for regeneration and selection of marker-free transgenic plants. In Vitro Cell Dev Biol Plant 37:103–113
Ebinuma H, Sugita K, Matsunaga E, Yamakado M (1997) Selection of marker-free transgenic plants using the isopentenyl transferase gene. Proc Natl Acad Sci 94:2117–2121
Endo S, Kasahara T, Sugita K, Matsunaga E, Ebinuma H (2001) The isopentenyl transferase gene is effective as a selectable marker gene for plant transformation in tobacco (Nicotiana tabacum cv. Petite Havana SRI). Plant Cell Rep 20:60–66
Endo S, Sugita K, Sakai M, Tanaka H, Ebinuma H (2002) Single-step transformation for generating marker-free transgenic rice using the ipt-type MAT vector system. Plant J 30:115–122
Ghorbel R, La-Malfa S, López MM, Petit A, Navarro L, Peña L (2001) Additional copies of virG from pTiBo542 provide a super-transformation ability to Agrobacterium tumefaciens in citrus. Physiol Mol Plant Pathol 58:103–110
Hake S, Smith HM, Holtan H, Magnani E, Mele G, Ramirez J (2004) The role of knox genes in plant development. Annu Rev Cell Dev Biol 20:125–151
Hay A, Tsiantis M (2010) KNOX genes: versatile regulators of plant development and diversity. Development 137:3153–3165
Khan EU, Fu XZ, Liu JH (2012) Agrobacterium-mediated genetic transformation and regeneration of transgenic plants using leaf segments as explants in Valencia sweet orange. Plant Cell Tissue Org Cult 109:383–390
Khawale RN, Singh SK, Garg G, Baranwal VK, Alizadeh Ajirlo S (2006) Agrobacterium-mediated genetic transformation of Nagpur mandarin (Citrus reticulata Blanco). Curr Sci 91:1700–1705
Kunkel T, Niu QW, Chan YS, Chua NH (1999) Inducible isopentenyl transferase as a high-efficiency marker for plant transformation. Nat Biotechnol 17:916–919
Li Y (2013) Gene deletor: a new tool to address gene flow and food safety concerns over transgenic crop plants. Front Biol 7:565–576
Li Y, Hagen G, Guilfoyle TJ (1991) An auxin-responsive promoter is differentially induced by auxin gradients during tropisms. Plant Cell 3:1167–1175
Li Y, Hagen G, Guilfoyle TJ (1992) Altered morphology in transgenic tobacco plants that overproduce cytokinins in specific tissues and organs. Dev Biol 153:386–395
López-Noguera S, Petri C, Burgos L (2009) Combining a regeneration-promoting ipt gene and site-specific recombination allows a more efficient apricot transformation and the elimination of marker genes. Plant Cell Rep 28:1781–1790
Luo K, Zheng X, Chen Y, Xiao Y, Zhao D, McAvoy R, Pei Y, Li Y (2006) The maize Knotted1 gene is an effective positive selectable marker gene for Agrobacterium-mediated tobacco transformation. Plant Cell Rep 25:403–409
Luo K, Duan H, Zhao D, Zheng X, Deng W, Chen Y, Stewart CNJ, McAvoy R, Jiang X, Wu Y, He A, Pei Y, Li Y (2007) ‘GM-gene-deletor’: fused loxP-FRT recognition sequences dramatically improve the efficiency of FLP or CRE recombinase on transgene excision from pollen and seed of tobacco plants. Plant Biotechnol J 5:263–274
Marutani-Hert M, Bowman KD, McCollum GT, Mirkov TE, Evens TJ, Niedz RP (2012) A dark incubation period is important for Agrobacterium-mediated transformation of mature internode explants of sweet orange, grapefruit, citron, and a citrange rootstock. PloS one 7:e47426
Mihálka V, Balazs E, Nagy I (2003) Binary transformation systems based on ‘shooter’ mutants of Agrobacterium tumefaciens: a simple, efficient and universal gene transfer technology that permits marker gene elimination. Plant Cell Rep 21:778–784
Moon J, Hake SC (2011) How a leaf gets its shape. Curr Opin Plant Biol 14:24–30
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497
Oliveira ML, Febres VJ, Costa MG, Moore GA, Otoni WC (2009) High-efficiency Agrobacterium-mediated transformation of citrus via sonication and vacuum infiltration. Plant Cell Rep 28:387–395
Orbović V, Dutt M, Grosser JW (2011) Seasonal effects of seed age on regeneration potential and transformation success rate in three citrus cultivars. Sci Hortic 127:262–266
Pei H, Zhao D, Song B (2010) Genetic transformation of explant with KN1 gene and its overexpression on transgenic plant in Jatropha curcas L. Genom Appl Biol 29:419–426
Pena L, Cervera M, Fagoaga C, Romero J, Juarez J, Pina JA, Navarro L (2007) Citrus. In: Nagata T (ed) Biotechnology in agriculture and forestry, vol 60. Springer, Berlin, pp 35–50
Peng AH, Xu LZ, He YR, Lei TG, Yao LX, Chen SC, Zou XP (2015) Efficient production of marker-free transgenic ‘Tarocco’ blood orange (Citrus sinensis Osbeck) with enhanced resistance to citrus canker using a Cre/loxP site-recombination system. Plant Cell, Tissue Organ Cult 123:1–13
Sander JD, Joung JK (2014) CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32:347–355
Sano R, Juarez CM, Hass B, Sakakibara K, Ito M, Banks JA, Hasebe M (2005) KNOX homeobox genes potentially have similar function in both diploid unicellular and multicellular meristems, but not in haploid meristems. Evol Dev 7:69–78
Semiarti E, Indrianto A, Pureantoro A, Isminingsih S, Suseno N, Ishikawa T, Yoshioka Y, Machida Y, Machida C (2007) Agrobacterium-mediated transformation of the wild orchid species Phalaenopsis amabilis. Plant Biotechnol 24:265–272
Sinha N, Williams R, Hake S (1993) Overexpression of the maize homeo box gene, KNOTTED-1, causes a switch from determinate to indeterminate cell fates. Genes Dev 7:787–795
Smigocki A, Owens L (1988) Cytokinin gene fused with a strong promoter enhances shoot organogenesis and zeatin levels in transformed plant cells. Proc Natl Acad Sci 85:5131–5135
Smigocki AC, Owens LD (1989) Cytokinin-to-auxin ratios and morphology of shoots and tissues transformed by a chimeric isopentenyl transferase gene. Plant Physiol 91:808–811
Strabala TJ, Bednarek SY, Bertoni G, Amasino RM (1989) Isolation and characterization of an ipt gene from the Ti plasmid Bo542. Mol Gen Genet 216:388–394
Strabala TJ, Wu YH, Li Y (1996) Combinatorial effects of cytokinin and auxin transport inhibitors: alteration of organogenesis and organ development from the shoot apical meristem. Plant Cell Physiol 37:1178–1182
Tamaoki M, Kusaba S, Kano-Murakami Y, Matsuoka M (1997) Ectopic expression of a tobacco homeobox gene, NTH15, dramatically alters leaf morphology and hormone levels in transgenic tobacco. Plant Cell Physiol 38:917–927
van Nocker S, Gardiner SE (2014) Breeding better cultivars, faster: applications of new technologies for the rapid deployment of superior horticultural tree crops. Hortic Res 1:22
Wang N, Trivedi P (2013) Citrus huanglongbing: a newly relevant disease presents unprecedented challenges. Phytopathology 103:652–665
Williams-Carrier RE, Lie YS, Hake S, Lemaux PG (1997) Ectopic expression of the maize kn1 gene phenocopies the hooded mutant of barley. Development 124:3737–3745
Xiao X, Ma F, Chen CL, Guo WW (2014) High efficient transformation of auxin reporter gene into trifoliate orange via Agrobacterium rhizogenes-mediated co-transformation. Plant Cell, Tissue Organ Cult 118:137–146
Xiong JS, Ding J, Li Y (2015) Genome-editing technologies and their potential application in horticultural crop breeding. Hortic Res 2:15019
Yan J, Yuan F, Long G, Qin L, Deng Z (2012) Selection of reference genes for quantitative real-time RT-PCR analysis in citrus. Mol Biol Rep 39:1831–1838
Yang L, Hu CH, Li N, Zhang JY, Yan JW, Deng ZN (2011) Transformation of sweet orange [Citrus sinensis (L.) Osbeck] with pthA-nls for acquiring resistance to citrus canker disease. Plant Mol Biol 75:11–23
Zheng X, Deng W, Luo K, Duan H, Chen Y, McAvoy R, Song S, Pei Y, Li Y (2007) The cauliflower mosaic virus (CaMV) 35S promoter sequence alters the level and patterns of activity of adjacent tissue- and organ-specific gene promoters. Plant Cell Rep 26:1195–1203
Acknowledgments
We are grateful to the Florida Citrus Research and Development Foundation for providing funds to support this project (Project Number 749). We are also thankful for Dr. Vladimir Orbovic of Citrus Research and Education Center at University of Florida for kindly providing us some of the citrus seeds used in this study. WH is a Ph.D. student at Hunan Agricultural University, Changsha, P. R. China. YL was a Shenlong Chair Visiting Professor of Hunan Agricultural University from 2012–2014.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Hu, W., Li, W., Xie, S. et al. Kn1 gene overexpression drastically improves genetic transformation efficiencies of citrus cultivars. Plant Cell Tiss Organ Cult 125, 81–91 (2016). https://doi.org/10.1007/s11240-015-0931-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-015-0931-z