Abstract
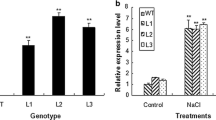
It has been over 50 years since the first prolyl aminopeptidase gene was identified in Escherichia coli (EC 3.4.11.5). However, up to now, few prolyl aminopeptidases have been reported to regulate osmotic stress tolerance, especially in plant. In this study, we focused on characterization of the biological functions of the Arabidopsis prolyl aminopeptidase AtPAP1 (At2g14260), which positively regulated plant tolerance to salt and drought stresses. Protein sequence alignment revealed that AtPAP1 was evolutionarily conserved among different plant species, and the smaller molecular weight and phylogenetic tree indicated that AtPAP1 belonged to the S33.001 subfamily. By using quantitative real-time PCR assays, we demonstrated that expression of the AtPAP1 gene was rapidly induced by salt and drought stresses. We also found that knockout of the AtPAP1 gene decreased, while AtPAP1 overexpression enhanced plant tolerance to salt and drought stresses. Measurements of the proline contents and the prolyl aminopeptidase activity suggested that the transgenic plants accumulated more free proline and exhibited higher prolyl aminopeptidase activity than the wild type or knockout plants under control conditions, as well as salt and drought stresses. Furthermore, through the GUS activity analysis, we also demonstrated that the AtPAP1 promoter is stress inducible and tissue specific. The AtPAP1-GFP fusion protein was found to localize in the cytoplasm of the onion epidermal cells. In conclusion, we showed that the Arabidopsis AtPAP1 gene could positively regulate plant tolerance to salt and drought stress, maybe by acting as a prolyl aminopeptidase and thereby increasing the concentration of free proline in plant cells.







Similar content being viewed by others
Abbreviations
- GUS:
-
β-Glucuronidase
- KO:
-
Knockout
- MU:
-
4-Methylumbelliferone
- MW:
-
Molecular weight
- OX:
-
Overexpression
- P5CR:
-
P5C reductase
- P5CS:
-
∆-1-Pyrroline-5-carboxylate synthetase
- PAP:
-
Prolyl aminopeptidase
- pI:
-
Isoelectric point
- PiP:
-
Proline iminopeptidase
- Pro-pNA:
-
Proline-p-nitroanilide trifluoroacetate
- ROS:
-
Reactive oxygen species
- WT:
-
Wild type
- X-Gluc:
-
5-Bromo-4-chloro-3-indolyl-β-d-glucuronide
References
Albertson NH, Koomey M (1997) Molecular cloning and characterization of a proline iminopeptidase from Serratia marcescens: cloning of the enzyme gene and crystallization of the expressed enzyme. J Biochem 122:601–605
Allaker RP, Young KA, Hardie JM (1994) Rapid detection of proline iminopeptidase as an indicator of Eikenella corrodens periodontal infection. Lett Appl Microbiol 19:325–327
Barnett NM, Naylor AW (1966) Amino acid and protein metabolism in Bermuda grass during water stress. Plant Physiol 41:1222–1230
Bates LS, Waldren RP, Teare ID (1973) Rapid determination of free proline in water-stress studies. Plant Soil 39:205–207
Bolumar T, Sanz Y, Aristory MC, Toldra F (2003) Purification and characterization of a prolyl aminopeptidase from Debaryomyces hansenii. Appl Environ Microbiol 69:227–232
Chliep MS, Ebert B, Simon-Rosin U, Zoeller D, Fisahn J (2010) Quantitative expression analysis of selected transcription factors in pavement, basal and trichome cells of mature leaves from Arabidopsis thaliana. Protoplasma 241:29–36
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cunningham DF, O’Connor B (1997) Proline specific peptidases. Biochim Biophys Acta 1343:160–186
Dixon DP, Skipsey M, Grundy NM, Edwards R (2005) Stress-induced protein and protein S-Glutathionylation in Arabidopsis. Plant Physiol 138:2233–2244
Dolferus R, Ji XM, Richards RA (2011) Abiotic stress and control of grain number in cereals. Plant Sci 181:331–341
FitzGerald RJ, O’Cuinn G (2006) Enzymatic debittering of food protein hydrolysates. Biotechnol Adv 24:234–237
Gao S, Yuan L, Zhai H, Liu CL, He SZ, Liu QC (2011) Transgenic sweetpotato plants expressing an LOS5 gene are tolerant to salt stress. Plant Cell Tiss Organ Cult 107:205–213
Gilbert C, Atlan D, Banc B, Portalier R (1994) Proline iminopeptidase from Lacto-bacillus delbrueckii subsp. bulgaricus CNRZ 397: purification and characterization. Microbiology 140:537–542
Islam SM, Tuteja N (2012) Enhancement of androgenesis by abiotic stress and other pretreatments in major crop species. Plant Sci 182:134–144
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Jin TC, Chang Q, Li WF, Yin DX, Li ZJ, Wang DL, Liu B, Liu LX (2010) Stress-inducible expression of GmDREB1 conferred salt tolerance in transgenic alfalfa. Plant Cell Tiss Organ Cult 100:219–227
Karthikeyan A, Pandian SK, Ramesh M (2011) Transgenic indica rice cv. ADT 43 expressing a ∆1-pyrroline-5-carboxylate synthetase (P5CS) gene from Vigna aconitifolia demonstrates salt tolerance. Plant Cell Tiss Organ Cult 107:383–395
Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50:347–363
Kishor P, Hong Z, Miao GH, Hu C, Verma D (1995) Overexpression of [delta]-pyrroline-5-carboxylate synthetase increases proline production and confers osmotolerance in transgenic plants. Plant Physiol 108:1387–1394
Kitazono A, Kitano A, Tsuru D, Yoshimoto T (1994) Isolation and characterization of the prolyl aminopeptidase gene (pap) from Aeromonas sobria: comparison with the Bacillus coagulans enzyme. J Biochem 116:818–825
Kitazono A, Kitano A, Kabashima T, Ito K, Yoshimoto T (1996) Prolyl aminopeptidase is also present in Enterobacteriaceae: cloning and sequencing of the Hafnia alvei enzyme-gene and characterization of the expressed enzyme. J Biochem 119:468–474
Krasensky J, Jonak C (2012) Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J Exp Bot 63:1593–1608
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9:299–306
Li N, Wu JM, Zhang LF, Zhang YZ, Feng H (2010) Characterization of a unique proline iminopeptidase from white-rot basidiomycetes Phanerochaete chrysosporium. Biochimie 92:779–788
Mahon S, O’Donoghue AJ, Goetz DH, Murray PG, Craik CS, Tuohy MG (2009) Characterization of a multimeric, eukaryotic prolyl aminopeptidase: an inducible and highly specific intracellular peptidase from the non-pathogenic fungus Talaromyces emersonii. Microbiology 155:3673–3682
Marinovaa M, Dolashkia A, Altenberend F, Stevanovic S, Voelter W, Tchorbanova B (2008) Characterization of an aminopeptidase and a proline iminopeptidase from cabbage leaves. Z Naturforsch C 63:105–112
Miller G, Honig A, Stein H, Suzuki N, Mittler R, Zilberstein A (2009) Unraveling delta1-pyrroline-5-carboxylate-proline cycle in plants by uncoupled expression of proline oxidation enzymes. J Biol Chem 284:26482–26492
Mohamed MA, Ibrahim TA (2012) Enhanced in vitro production of Ruta graveolens L. coumarins and rutin by mannitol and ventilation. Plant Cell Tiss Organ Cult 111:335–343
Mueller LA, Hinz U, Uzé M, Sautter C, Zryd JP (1997) Biochemical complementation of the betalain biosynthetic pathway in Portulaca grandiflora by a fungal 3,4-dihydroxyphenylalanine dioxygenase. Planta 203:260–263
Nishiuchi T, Hamada T, Kodama H, Iba K (1997) Wounding changes the spatial expression pattern of the Arabidopsis plastid omega-3 fatty acid desaturase gene (FAD7) through different signal transduction pathways. Plant Cell 9:1701–1712
Oviedo Ovando ME, Isola MC, Maldonado AM, Franzoni L (2004) Purification and properties of iminopeptidase from peanut seeds. Plant Sci 166:1143–1148
Patade VY, Bhargava S, Suprasanna P (2012) Effects of NaCl and iso-osmotic PEG stress on growth, osmolytes accumulation and antioxidant defense in cultured sugarcane cells. Plant Cell Tiss Organ Cult 108:279–286
Peng ZH, Peng KQ, Hu JJ, Xiao LT (2002) Reseach progress on accumulation of proline under osmotic stress in plants. Chin Agric Sci Bull 18:80–83
Qu LJ, Wu LQ, Fan ZM, Guo L, Li YQ, Chen ZL (2005) Over-expression of the bacterial nhaA gene in rice enhances salt and drought tolerance. Plant Sci 168:297–302
Quan XQ, Zhang YJ, Shan L, Bi YP (2007) The roles of prolines in plant growth and the tolerance to abiotic stresses. Lett Biotechnol 18:159–162
Sarid S, Berger A, Katchalski E (1959) Proline iminopeptide. J Biol Chem 234:1740–1746
Selby T, Allaker RP, Dymock D (2003) Characterization and expression of adjacent proline iminopeptidase and aspartase genes from Eikenella corrodens. Oral Microbiol Immunol 18:256–259
Shangguan XX, Xu B, Yu ZX, Wang LJ, Chen XY (2008) Promoter of a cotton fibre MYB gene functional in trichomes of Arabidopsis and glandular trichomes of tobacco. J Exp Bot 59:3533–3542
Somboonwatthanaku I, Dorling S, Leung S, McManus MT (2010) Proline biosynthetic gene expression in tissue cultures of rice (Oryza sativa L.) in response to saline treatment. Plant Cell Tiss Organ Cult 103:369–376
Stein H, Honig A (2011) Elevation of free proline and proline-rich protein levels by simultaneous manipulations of proline biosynthesis and degradation in plants. Plant Sci 181:140–150
Stewart GR, Lee JA (1974) Role of proline accumulation in halophytes. Planta 120:279–289
Szabados L, Savoure A (2010) Proline: a multifunctional amino acid. Trends Plant Sci 15:89–97
Szawłowska U, Zdunek-Zastocka E, Bielawski W (2011) Biochemical characterisation of prolyl aminopeptidase from shoots of triticale seedlings and its activity changes in response to suboptimal growth conditions. Plant Physiol Biochem 49:1342–1349
Szawłowska U, Grabowska A, Zdunek-Zastocka E, Bielawski W (2012) TsPAP1 encodes a novel plant prolyl aminopeptidase whose expression is induced in response to suboptimal growth conditions. Biochem Biophys Res Commun 419:104–109
Tocquin P, Corbesier L, Havelange A, Pieltain A, Kurtem E, Bernier G, Périlleux C (2003) A novel high efficiency, low maintenance, hydroponic system for synchronous growth and flowering of Arabidopsis thaliana. BMC Plant Biol 3:2
Weterings K, Schrauwen J, Wullems G, Twell D (1995) Functional dissection of the promoter of the pollen-specific gene NPT303 reveals a novel pollen-specific, and conserved cis-regulatory element. Plant J 8:55–63
Willems E, Leyns L, Vandesompele J (2008) Standardization of real-time PCR gene expression data from independent biological replicates. Anal Biochem 379:127–129
Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An ‘electronic fluorescent pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2:e718
Yang YL, Yang F, Li XN, Shi RX, Lu J (2013) Signal regulation of proline metabolism in callus of the halophyte Nitraria tangutorum Bobr. grown under salinity stress. Plant Cell Tiss Organ Cult 112:33–42
Yoshimoto T, Tsuru D (1985) Proline iminopeptidase from Bacillus coagulans: purification and enzymatic properties. J Biochem 97:1477–1485
Zhang L, Jia Y, Wang L, Wang R (2007) A proline iminopeptidase gene upregulated in planta by a LuxR homologue is essential for pathogenicity of Xanthomonas campestris pv, campestris. Mol Microbiol 65:121–136
Acknowledgments
This work was financially supported by Heilongjiang Provincial Higher School Science and Technology Innovation Team Building Program (2011TD005), National Natural Science Foundation of China (31171578) and the Heilongjiang Provincial Graduate Student Innovation Research Projects (YJSCX2012-047HLJ).
Author information
Authors and Affiliations
Corresponding author
Additional information
Xiaoli Sun and Feifei Wang equally contributed to this work.
Rights and permissions
About this article
Cite this article
Sun, X., Wang, F., Cai, H. et al. Functional characterization of an Arabidopsis prolyl aminopeptidase AtPAP1 in response to salt and drought stresses. Plant Cell Tiss Organ Cult 114, 325–338 (2013). https://doi.org/10.1007/s11240-013-0328-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-013-0328-9