Abstract
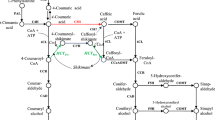
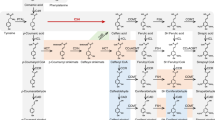
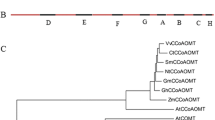
Studying lignin biosynthesis in Panicum virgatum (switchgrass) has provided a basis for generating plants with reduced lignin content and increased saccharification efficiency. Chlorogenic acid (CGA, caffeoyl quinate) is the major soluble phenolic compound in switchgrass, and the lignin and CGA biosynthetic pathways potentially share intermediates and enzymes. The enzyme hydroxycinnamoyl-CoA: quinate hydroxycinnamoyltransferase (HQT) is responsible for CGA biosynthesis in tobacco, tomato and globe artichoke, but there are no close orthologs of HQT in switchgrass or in other monocotyledonous plants with complete genome sequences. We examined available transcriptomic databases for genes encoding enzymes potentially involved in CGA biosynthesis in switchgrass. The protein products of two hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyltransferase (HCT) genes (PvHCT1a and PvHCT2a), closely related to lignin pathway HCTs from other species, were characterized biochemically and exhibited the expected HCT activity, preferring shikimic acid as acyl acceptor. We also characterized two switchgrass coumaroyl shikimate 3′-hydroxylase (C3′H) enzymes (PvC3′H1 and PvC3′H2); both of these cytochrome P450s had the capacity to hydroxylate 4-coumaroyl shikimate or 4-coumaroyl quinate to generate caffeoyl shikimate or CGA. Another switchgrass hydroxycinnamoyl transferase, PvHCT-Like1, is phylogenetically distant from HCTs or HQTs, but exhibits HQT activity, preferring quinic acid as acyl acceptor, and could therefore function in CGA biosynthesis. The biochemical features of the recombinant enzymes, the presence of the corresponding activities in plant protein extracts, and the expression patterns of the corresponding genes, suggest preferred routes to CGA in switchgrass.







Similar content being viewed by others
References
Ahola V, Aittokallio T, Vihinen M, Uusipaikka E (2006) A statistical score for assessing the quality of multiple sequence alignments. BMC Bioinformat 7:484
Chen F, Dixon RA (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25:759–761
Chen F, Srinivasa Reddy MS, Temple S, Jackson L, Shadle G, Dixon RA (2006) Multi-site genetic modulation of monolignol biosynthesis suggests new routes for formation of syringyl lignin and wall-bound ferulic acid in alfalfa (Medicago sativa L.). Plant J 48:113–124
Comino C, Lanteri S, Portis E, Acquadro A, Romani A, Hehn A, Larbat R, Bourgaud F (2007) Isolation and functional characterization of a cDNA coding a hydroxycinnamoyltransferase involved in phenylpropanoid biosynthesis in Cynara cardunculus L. BMC Plant Biol 7:14
Comino C, Hehn A, Moglia A, Menin B, Bourgaud F, Lanteri S, Portis E (2009) The isolation and mapping of a novel hydroxycinnamoyltransferase in the globe artichoke chlorogenic acid pathway. BMC Plant Biol 9:30
Davison BH, Drescher SR, Tuskan GA, Davis MF, Nghiem NP (2006) Variation of S/G ratio and lignin content in a Populus family influences the release of xylose by dilute acid hydrolysis. Appl Biochem Biotechnol 129–132:427–435
Escamilla-Trevino LL, Shen H, Uppalapati SR, Ray T, Tang Y, Hernandez T, Yin Y, Xu Y, Dixon RA (2010) Switchgrass (Panicum virgatum) possesses a divergent family of cinnamoyl CoA reductases with distinct biochemical properties. New Phytol 185:143–155
Fu C, Mielenz JR, Xiao X, Ge Y, Hamilton CY, Rodriguez M Jr, Chen F, Foston M, Ragauskas A, Bouton J, Dixon RA, Wang Z-Y (2011) Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc Natl Acad Sci USA 108:3803–3808
Gallego-Giraldo L, Escamilla-Trevino L, Jackson LA, Dixon RA (2011) Salicylic acid mediates the reduced growth of lignin down-regulated plants. Proc Natl Acad Sci USA 108:20814–20819
Grabber JH (2005) How do lignin composition, structure, and cross-linking affect degradability? A review of cell wall model studies. Crop Sci 45:820–831
Hoffmann L, Maury S, Martz F, Geoffroy P, Legrand M (2003) Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J Biol Chem 278:95–103
Hoffmann L, Besseau S, Geoffroy P, Ritzenthaler C, Meyer D, Lapierre C, Pollet B, Legrand M (2004) Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16:1446–1465
Jackson LA, Shadle GL, Zhou R, Nakashima J, Chen F, Dixon RA (2008) Improving saccharification efficiency of alfalfa stems through modification of the terminal stages of monolignol biosynthesis. Bioenergy Res 1:180–192
Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518
Lepelley M, Cheminade G, Tremillon N, Simkin A, Caillet V, McCarthy J (2007) Chlorogenic acid synthesis in coffee: an analysis of CGA content and real-time RT-PCR expression of HCT, HQT, C3H1, and CCoAOMT1 genes during grain development in C. canephora. Plant Sci 172:978–996
Liu CJ, Huhman D, Sumner LW, Dixon RA (2003) Regiospecific hydroxylation of isoflavones by cytochrome P450 81E enzymes from Medicago truncatula. Plant J 36:471–484
Mahesh V, Million-Rousseau R, Ullmann P, Chabrillange N, Bustamante J, Mondolot L, Morant M, Noirot M, Hamon S, de Kochko A (2007) Functional characterization of two p-coumaroyl ester 3′-hydroxylase genes from coffee tree: evidence of a candidate for chlorogenic acid biosynthesis. Plant Mol Biol 64:145–159
McLaughlin SB, Adams Kszos L (2005) Development of switchgrass (Panicum virgatum) as a bioenergy feedstock in the United States. Biomass Bioenergy 28:515–535
Moglia A, Comino C, Portis E, Acquadro A, De Vos RCH, Beekwilder J, Lanteri S (2009) Isolation and mapping of a C3′H gene (CYP98A49) from globe artichoke, and its expression upon UV–C stress. Plant Cell Rep 28:963–974
Moore KJ, Moser LE, Vogel KP, Waller SS, Johnson BE, Pedersen JF (1991) Describing and quantifying growth stages of perennial forage grasses. Agron J 83:1073–1077
Morant M, Schoch GA, Ullmann P, Ertunҫ T, Little D, Olsen CE, Petersen M, Negrel J, Werck-Reichhart D (2007) Catalytic activity, duplication and evolution of the CYP98 cytochrome P450 family in wheat. Plant Mol Biol 63:1–19
Niggeweg R, Michael AJ, Martin C (2004) Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat Biotechnol 22:746–754
Nuin PA, Wang Z, Tillier ER (2006) The accuracy of several multiple sequence alignment programs for proteins. BMC Bioinformat 7:471
Pompon D, Louerat B, Bronine A, Urban P (1996) Yeast expression of animal and plant P450 s in optimized redox environments. Meth Enzymol 272:51–64
Price MN, Dehal PS, Arkin AP (2010) FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490
Ralph J, Akiyama T, Kim H, Lu F, Schatz PF, Marita JM, Ralph SA, Reddy MS, Chen F, Dixon RA (2006) Effects of coumarate 3′-hydroxylase down-regulation on lignin structure. J Biol Chem 281:8843–8853
Reddy MS, Chen F, Shadle G, Jackson L, Aljoe H, Dixon RA (2005) Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.). Proc Natl Acad Sci USA 102:16573–16578
Schmer MR, Vogel KP, Mitchell RB, Perrin RK (2008) Net energy of cellulosic ethanol from switchgrass. Proc Natl Acad Sci USA 105:464–469
Schoch G, Goepfert S, Morant M, Hehn A, Meyer D, Ullmann P, Werck-Reichhart D (2001) CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J Biol Chem 276:36566–36574
Shen H, Fu CX, Xiao XR, Ray T, Tang YH, Wang ZY, Chen F (2009) Developmental control of lignification in stems of lowland switchgrass variety Alamo and the effects on saccharification efficiency. Bioenergy Res 2:233–245
Shen H, He X, Poovaiah CR, Wuddineh WA, Ma J, Mann DGJ, Wang H, Jackson L, Tang Y, Neal Stewart C Jr (2011) Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol 193:121–136
Shen H, Mazarei M, Rudis MR, Tang YH, Hisano H, Jackson L, Li G, Hernandez T, Chen F, C. Stewart Jr N, Wang Z, Dixon RA (2013). A genomic approach to deciphering pathways for lignin biosynthesis in switchgrass (Panicum virgatum L.). Plant Cell, in revision
Sonnante G, D’Amore R, Blanco E, Pierri CL, De Palma M, Luo J, Tucci M, Martin C (2010) Novel hydroxycinnamoyl-coenzyme A quinate transferase genes from artichoke are involved in the synthesis of chlorogenic acid. Plant Physiol 153:1224–1238
Stockigt J, Zenk MH (1975) Chemical syntheses and properties of hydroxycinnamoyl-Coenzyme A derivatives. Z Naturforsch [C] 30:352–358
Talukder K (2006) Low-lignin wood–a case study. Nat Biotechnol 24:395–396
van der Rest B, Danoun S, Boudet AM, Rochange SF (2006) Down-regulation of cinnamoyl-CoA reductase in tomato (Solanum lycopersicum L.) induces dramatic changes in soluble phenolic pools. J Exp Bot 57:1399–1411
Vanholme R, Cesarino I, Rataj K, Xiao Y, Sundin L, Goeminne G, Kim H, Cross J, Morreel K, Araujo P, Welsh L, Haustraete J, McClellan C, Vanholme B, Ralph J, Simpson GG, Halpin C, Boerjan W (2013) Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway. Science 341:1103–1106
Villegas RJ, Kojima M (1986) Purification and characterization of hydroxycinnamoyl d-glucose: quinate hydroxycinnamoyl transferase in the root of sweet potato, Ipomoea batatas Lam. J Biol Chem 261:8729–8733
Wullschleger SD, Davis EB, Borsuk ME, Gunderson CA, Lynd LR (2010) Biomass production in switchgrass across the United States: database description and determinants of yield. Agron J 102:1158–1168
Xu B, Escamilla-Trevino LL, Noppadon S, Shen Z, Shen H, Percival Zhang YH, Dixon RA, Zhao B (2011) Silencing of 4-coumarate: coenzyme A ligase in switchgrass leads to reduced lignin content and improved fermentable sugar yields for biofuel production. New Phytol 92:611–625
Yuan JS, Tiller KH, Al-Ahmad H, Stewart NR, Stewart CN (2008) Plants to power: bioenergy to fuel the future. Trends Plant Sci 13:421–429
Zhang J-Y, Lee Y-C, Torres-Jerez I, Wang M, Yin Y, Chou W-C, He J, Shen H, Srivastava AC, Pennacchio C, Lindquist E, Grimwood J, Schmutz J, Xu Y, Sharma M, Sharma R, Bartley LE, Ronald PC, Saha MC, Dixon RA, Tang Y, Udvardi MK (2013) Development of an integrated transcript sequence database and a gene expression atlas for gene discovery and analysis in switchgrass (Panicum virgatum L.). Plant J 74:160–173
Acknowledgments
We thank Drs. Jerome Verdier and Lina Gallego-Giraldo for critical reading of the manuscript. This work was supported by the BioEnergy Science Center, a US Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science.
Author information
Authors and Affiliations
Corresponding author
Additional information
Accessions numbers
PvHCT1a: AB723827, PvHCT2a: KC696573, PvHCT-Like1: JX845714, PvC3′H1: AB723823, PvC3′H2: AB723824.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Escamilla-Treviño, L.L., Shen, H., Hernandez, T. et al. Early lignin pathway enzymes and routes to chlorogenic acid in switchgrass (Panicum virgatum L.). Plant Mol Biol 84, 565–576 (2014). https://doi.org/10.1007/s11103-013-0152-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-013-0152-y