Abstract
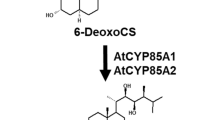
Cytochrome P450 monooxygenases (P450s) are a diverse family of proteins that have specialized roles in secondary metabolism and in normal cell development. Two P450s in particular, CYP734A1 and CYP72C1, have been identified as brassinosteroid-inactivating enzymes important for steroid-mediated signal transduction in Arabidopsis thaliana. Genetic analyses have demonstrated that these P450s modulate growth throughout plant development. While members of the CYP734A subfamily inactivate brassinosteroids through C-26 hydroxylation, the biochemical activity of CYP72C1 is unknown. Because CYP734A1 and CYP72C1 in Arabidopsis diverge more than brassinosteroid inactivating P450s in other plants, this study examines the structure and biochemistry of each enzyme. Three-dimensional models were generated to examine the substrate binding site structures and determine how they might affect the function of each P450. These models have indicated that the active site of CYP72C1 does not contain several conserved amino acids typically needed for substrate hydroxylation. Heterologous expression of these P450s followed by substrate binding analyses have indicated that CYP734A1 binds active brassinosteroids, brassinolide and castasterone, as well as their upstream precursors whereas CYP72C1 binds precursors more effectively. Seedling growth assays have demonstrated that the genetic state of CYP734A1, but not CYP72C1, affected responsiveness to high levels of exogenous brassinolide supporting our observations that CYP72C1 acts on brassinolide precursors. Although there may be some overlap in their physiological function, the distinct biochemical functions of these proteins in Arabidopsis has significant potential to fine-tune the levels of different brassinosteroid hormones throughout plant growth and development.







Similar content being viewed by others
Abbreviations
- P450:
-
Cytochrome P450
- BR:
-
Brassinosteroid
- SRS:
-
Substrate recognition site
References
Altmann T (1999) Molecular physiology of brassinosteroids revealed by the analysis of mutants. Planta 208:1–11
Anuradha S, Rao S (2001) Effect of brassinosteroids on salinity stress induced inhibition of seed germination and seedling growth in rice (Oryza sativa L.). Plant Growth Regul 33:151–153
Barnes HJ (1992) Maximizing expression of eukaryotic cytochrome P450s in Escherichia coli. Methods Enzymol 272:3–14
Barnes WM (1994) PCR amplification of up to 35 kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc Natl Acad Sci USA 91:2216–2220
Baudry J, Rupasinghe S, Schuler MA (2006) Class-dependent sequence alignment strategy improves the structural and functional modeling of P450s. Protein Eng Des Sel 19:345–353
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235–242
Bernhardt R, Waterman MR (2007) Cytochrome P450 and steroid hormone biosynthesis. In: Sigel A, Sigel H, Sigel RKO (eds) The ubiquitous roles of P450 proteins, vol 3. Wiley, Chichester, pp 361–396
Choe S (2007) Signal-transduction pathways toward the regulation of brassinosteroid biosynthesis. J Plant Biol 50:225–229
Duan H, Civjan NR, Sligar SG, Schuler MA (2004) Co-incorporation of heterologously expressed Arabidopsis cytochrome P450 and P450 reductase into soluble nanoscale lipid bilayers. Arch Biochem Biophys 424:141–153
Fujioka S, Yokota T (2003) Biosynthesis and metabolism of brassinosteroids. Ann Rev Plant Biol 54:137–164
Gegner JA, Dahlquist FW (1991) Signal transduction in bacteria: CheW forms a reversible complex with the protein kinase CheA. Proc Natl Acad Sci USA 88:750–754
Gillam EMJ, Hunter DJB (2007) Chemical defense and exploitation. Biotransformation of xenobiotics by cytochrome P450 enzymes. In: Sigel A, Sigel H, Sigel RKO (eds) The ubiquitous roles of P450 proteins, vol 3. Wiley, Chichester, pp 477–560
Gotoh O (1992) Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem 267:83–90
Graham SE, Peterson JA (1999) How similar are P450s and what can their differences teach us? Arch Biochem Biophys 369:24–29
Halgren TA (1996) Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem 17:490–519
Jefcoate CR (1978) Measurement of substrate and inhibitor binding to microsomal cytochrome P-450 by optical-difference spectroscopy. Methods Enzymol 52:258–279
Johnson EF, Stout CD (2005) Structural diversity of human xenobiotic-metabolizing cytochrome P450 monooxygenases. Biochem Biophys Res Commun 338:331–336
Kemp CA, Marechal JD, Sutcliffe MJ (2005) Progress in cytochrome P450 active site modeling. Arch Biochem Biophys 433:361–368
Kim T-W, Wang Z-Y (2010) Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol 61:681–704
Kim TW, Hwang J-Y, Kim Y-S, Joo S-H, Chang SC, Lee JS, Takatsuto S, Kim S-K (2005) Arabidopsis CYP85A2, a cytochrome P450, mediates the Baeyer–Villiger oxidation of castasterone to brassinolide in brassinosteroid biosynthesis. Plant Cell 17:2397–2412
Lee DS, Yamada A, Sugimoto H, Matsunaga I, Ogura H, Ichihara K, Adachi S, Park SY, Shiro Y (2003) Substrate recognition and molecular mechanism of fatty acid hydroxylation by cytochrome P450 from Bacillus subtilis. Crystallographic, spectroscopic, and mutational studies. J Biol Chem 278:9761–9767
Lee D-S, Nioche P, Hamberg M, Raman CS (2008) Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes. Nature 455:363–368
Lewis DF (2004) 57 varieties: the human cytochromes P450. Pharmacogenomics 5:305–318
Leys D, Mowat CG, McLean KJ, Richmond A, Chapman SK, Walkinshaw MD, Munro AW (2003) Atomic structure of Mycobacterium tuberculosis CYP121 to 1.06 A reveals novel features of cytochrome P450. J Biol Chem 278:5141–5147
Li J, Chory J (1999) Brassinosteroid actions in plants. J Exp Bot 50:272–282
MacKerell AD Jr, Bashford D, Bellott M, Dunbrack RL Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102:3585–3616
Makris TM, Denisov I, Schlichting I, Sligar S (2005) Activation of molecular oxygen by cytochrome P450. In: Ortiz de Montellano PR (ed) Cytochrome P450: structure, mechanism, and biochemistry, 3rd edn. Kluwer Academic/Plenum Publishers, New York, pp 149–182
Marsolais F, Boyd J, Paredes Y, Schinas AM, Garcia M, Elzein S, Varin L (2007) Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a (At2g14920) and AtST1 (At2g03760). Planta 225:1233–1244
Nagano S, Li H, Shimizu H, Nishida C, Ogura H, Ortiz de Montellano PR, Poulos TL (2003) Crystal structures of epothilone D-bound, epothilone B-bound, and substrate-free forms of cytochrome P450epoK. J Biol Chem 278:44886–44893
Nakamura M, Satoh T, Tanaka SI, Mochizuki N, Yokota T, Nagatani A (2005) Activation of the cytochrome P450 gene, CYP72C1, reduces the levels of active brassinosteroids in vivo. J Exp Bot 56:833–840
Nakashita H, Yasuda M, Nitta T, Asami T, Fujioka S, Arai Y, Sekimata K, Takatsuto S, Yamaguchi I, Yoshida S (2003) Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J 33:887–898
Neff MM, Nguyen SM, Malancharuvil EJ, Fujioka S, Noguchi T, Seto H, Tsubuki M, Honda T, Takatsuto S, Yoshida S, Chory J (1999) BAS1: a gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci USA 96:15316–15323
Nelson DR, Schuler MA, Paquette SM, Werck-Reichhart D, Bak S (2004) Comparative genomics of rice and Arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiol 135:756–772
Nelson DR, Ming R, Alam M, Schuler MA (2008) Comparison of cytochrome P450 genes from six plant genomes. Trop Plant Biol 1:216–235
Ohnishi T, Nomura T, Watanabe B, Ohta D, Yokota T, Miyagawa H, Sakata K, Mizutani M (2006a) Tomato cytochrome P450 CYP734A7 functions in brassinosteroid catabolism. Phytochemistry 67:1895–1906
Ohnishi T, Szatmari A-M, Watanabe B, Fujita S, Bancos S, Koncz C, Lafos M, Shibata K, Yokota T, Sakata K, Szekeres M, Mizutani M (2006b) C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell 18:3275–3288
Ohnishi T, Yokota T, Mizutani M (2009) Insights into the function and evolution of P450s in plant steroid metabolism. Phytochemistry 70:1918–1929
Omura T, Sato R (1964) The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem 239:2370–2378
Ortiz de Montellano PR (2005) Cytochrome P450: structure, mechanism, and biochemistry, 3rd edn. Kluwer Academic/Plenum Publishers, New York
Paquette SM, Bak S, Feyereisen R (2000) Intron-exon organization and phylogeny in a large superfamily, the paralogous cytochrome P450 genes of Arabidopsis thaliana. DNA Cell Biol 19:307–317
Podust LM, Yermalitskaya LV, Lepesheva GI, Podust VN, Dalmasso EA, Waterman MR (2004) Estriol bound and ligand-free structures of sterol 14alpha-demethylase. Structure 12:1937–1945
Poppenberger B, Fujioka S, Soeno K, George GL, Vaistij FE, Hiranuma S, Seto H, Takatsuto S, Adam G, Yoshida S, Bowles D (2005) The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc Natl Acad Sci USA 102:15253–15258
Poulos TL, Johnson EF (2005) Structures of cytochrome P450 enzymes. In: Ortiz de Montellano PR (ed) Cytochrome P450: structure, mechanism, and biochemistry, 3rd edn. Kluwer Academic/Plenum, New York, pp 87–114
Poulos TL, Meharenna YT (2007) Structures of P450 proteins and their molecular phylogenies. In: Sigel A, Sigel H, Sigel RKO (eds) The ubiquitous roles of P450 proteins, vol 3. Wiley, Chichester, pp 57–96
Poulos TL, Finzel BC, Howard AJ (1987) High-resolution crystal structure of cytochrome P450cam. J Mol Biol 195:687–700
Ravichandran KG, Boddupalli SS, Hasermann CA, Peterson JA, Deisenhofer J (1993) Crystal structure of hemoprotein domain of P450BM-3, a prototype for microsomal P450’s. Science 261:731–736
Rupasinghe S, Schuler MA (2006) Homology modeling of plant cytochrome P450s. Phytochem Rev 5:473–505
Rupasinghe SG, Duan H, Schuler MA (2007) Molecular definitions of fatty acid hydroxylases in Arabidopsis thaliana. Proteins 68:279–293
Schoch GA, Yano JK, Wester MR, Griffin KJ, Stout CD, Johnson EF (2004) Structure of human microsomal cytochrome P450 2C8. Evidence for a peripheral fatty acid binding site. J Biol Chem 279:9497–9503
Schuler MA, Werck-Reichhart D (2003) Functional genomics of P450s. Annu Rev Plant Biol 54:629–667
Scott EE, White MA, He YA, Johnson EF, Stout CD, Halpert JR (2004) Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9-A resolution: insight into the range of P450 conformations and the coordination of redox partner binding. J Biol Chem 279:27294–27301
Sigel A, Sigel H, Sigel RKO (2007) The ubiquitous roles of cytochrome P450 proteins, vol 3. Wiley, Chichester
Szklarz GD, Graham SE, Paulsen MD (2000) Molecular modeling of mammalian cytochromes P450: application to study enzyme function. Vitam Horm 58:53–87
Takahashi N, Nakazawa M, Shibata K, Yokota T, Ishikawa A, Suzuki K, Kawashima M, Ichikawa T, Shimada H, Matsui M (2005) shk1-D, a dwarf Arabidopsis mutant caused by activation of the CYP72C1 gene, has altered brassinosteroid levels. Plant J 42:13–22
Turk EM, Fujioka S, Seto H, Shimada Y, Takatsuto S, Yoshida S, Denzel MA, Torres QI, Neff MM (2003) CYP72B1 inactivates brassinosteroid hormones: an intersection between photomorphogenesis and plant steroid signal transduction. Plant Physiol 133:1643–1653
Turk EM, Fujioka S, Seto H, Shimada Y, Takatsuto S, Yoshida S, Wang H, Torres QI, Ward JM, Murthy G, Zhang J, Walker JC, Neff MM (2005) BAS1 and SOB7 act redundantly to modulate Arabidopsis photomorphogenesis via unique brassinosteroid inactivation mechanisms. Plant J 42:23–34
Wade RC, Winn PJ, Schlichting I, Sudarko (2004) A survey of active site access channels in cytochromes P450. J Inorg Biochem 98:1175–1182
Werck-Reichhart D, Bak S, Paquette S (2002) Cytochromes P450. In: CR Somerville, EM Meyerowitz (eds) The Arabidopsis book. American Society of Plant Biologists, Rockville, pp 1–28. doi:10.1199/tab.0028. http://www.aspb.org/publications/arabidopsis/. Accessed July 1, 2009
Wester MR, Johnson EF, Marques-Soares C, Dansette PM, Mansuy D, Stout CD (2003) Structure of a substrate complex of mammalian cytochrome P450 2C5 at 2.3 a resolution: evidence for multiple substrate binding modes. Biochemistry 42:6370–6379
Williams PA, Cosme J, Ward A, Angove HC, Matak Vinkovic D, Jhoti H (2003) Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature 424:464–468
Yano JK, Blasco F, Li H, Schmid RD, Henne A, Poulos TL (2003) Preliminary characterization and crystal structure of a thermostable cytochrome P450 from Thermus thermophilus. J Biol Chem 278:608–616
Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF (2004) The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-A resolution. J Biol Chem 279:38091–38094
Yuan T, Fujioka S, Takatsuto S, Matsumoto S, Xiaoping G, He K, Russell SD, Li J (2007) BEN1, a gene encoding a dihydroflavonol 4-reductase (DFR)-like protein, regulates the levels of brassinosteroids in Arabidopsis thaliana. Plant J 51:220–233
Zhao B, Guengerich FP, Bellamine A, Lamb DC, Izumikawa M, Lei L, Podust LM, Sundaramoorthy M, Kalaitzis JA, Reddy LM, Kelly SL, Moore BS, Stec D, Voehler M, Falck JR, Shimada T, Waterman MR (2005) Binding of two flaviolin substrate molecules, oxidative coupling, and crystal structure of Streptomyces coelicolor A3(2) cytochrome P450 158A2. J Biol Chem 280:11599–11607
Acknowledgments
We thank Dr. Zhimou Wen for technical input on the baculovirus expression and type-I binding experiments. We also thank Dr. Shozo Fujioka for his gift of castasterone precursors for use as binding substrates and Dr. Edward Turk for cloning the Arabidopsis CYP734A1 and CYP72C1 cDNAs. We thank Drs. Christopher Mau and Rodney Croteau for their help in establishing the E. coli expression system. This research was supported by the United States Department of Agriculture 2005-35318-16214 (L.E.T.), the National Science Foundation 0758411 (M.M.N.) and MCB0115068 (M.A.S.). We are also grateful for support from the Department of Energy DE-FG02-08ER15927 (M.M.N.).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Thornton, L.E., Rupasinghe, S.G., Peng, H. et al. Arabidopsis CYP72C1 is an atypical cytochrome P450 that inactivates brassinosteroids. Plant Mol Biol 74, 167–181 (2010). https://doi.org/10.1007/s11103-010-9663-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-010-9663-y