Abstract
Iron uptake and translocation in plants are important processes for both plant and human nutrition, whereas relatively little is known about the molecular mechanisms of iron transport within the plant body. Several reports have shown that yellow stripe 1 (YS1) and YS1-like (YSL) transporters mediate metal-phytosiderophore uptake and/or metal-nicotianamine translocation. Among the 18 YSL genes in rice (OsYSLs), OsYSL18 is predicted to encode a polypeptide of 679 amino acids containing 13 putative transmembrane domains. An OsYSL18-green fluorescent protein (GFP) fusion was localized to the plasma membrane when transiently expressed in onion epidermal cells. Electrophysiological measurements using Xenopus laevis oocytes showed that OsYSL18 transports iron(III)–deoxymugineic acid, but not iron(II)–nicotianamine, zinc(II)–deoxymugineic acid, or zinc(II)–nicotianamine. Reverse transcriptase PCR analysis revealed more OsYSL18 transcripts in flowers than in shoots or roots. OsYSL18 promoter-β-glucuronidase (GUS) analysis revealed that OsYSL18 was expressed in reproductive organs including the pollen tube. In vegetative organs, OsYSL18 was specifically expressed in lamina joints, the inner cortex of crown roots, and phloem parenchyma and companion cells at the basal part of every leaf sheath. These results suggest that OsYSL18 is an iron-phytosiderophore transporter involved in the translocation of iron in reproductive organs and phloem in joints.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The most widespread human nutritional problem is iron (Fe) deficiency. There are approximately two billion anemic people worldwide, and approximately 50% of all anemia’s can be attributed to Fe deficiency (Mason et al. 2001). Because plants are the primary source of food for humans, the nutritional value of plants is of central importance to human health (Grusak and Dellapenna 1999). Identification of plant mechanisms involved in the acquisition and use of Fe is a prerequisite to increasing the amount of Fe in the edible parts of plants. Identifying the transporters involved in uptake and translocation of Fe is an important aspect of this task.
Plants have evolved two distinct Fe uptake strategies (Römheld and Marschner 1986). Most plants, including dicots and non-graminaceous monocots, take up Fe(II) from soils into root cells via iron-regulated transporters (IRT) after reduction from ferric [Fe(III)]-chelates by ferric reductase/oxidase (FRO) in the plasma membrane (Strategy I; Eide et al. 1996; Robinson et al. 1999). However, graminaceous plants synthesize and secrete natural Fe chelators, mugineic acid family phytosiderophores (MAs), from their roots to solubilize Fe in the rhizosphere (Strategy II; Takagi 1976, Römheld and Marchner 1986; Mori 1999). The resulting Fe(III)–MAs complexes are absorbed into the root through Fe(III)–MAs transporters in the plasma membrane. The gene encoding an Fe(III)–MAs transporter, Yellow Stripe 1 (YS1), was first isolated in Zea mays (Curie et al. 2001). Electrophysiological analyses revealed that YS1 functions as a proton-coupled symporter for various MAs-bound metals including Fe(III), Zn(II), Cu(II), and Ni(II) (Schaff et al. 2004). Recently, a barley homolog of YS1 (HvYS1) was isolated and was shown to transport Fe(III)–MAs (Murata et al. 2006).
MAs are biosynthesized from methionine (Mori and Nishizawa 1987) via several enzymatic reactions (Shojima et al. 1990; Mori 1999; Bashir et al. 2006). Nicotianamine (NA), the biosynthetic precursor of MAs (Shojima et al. 1989, 1990), is structurally very similar to MAs and chelates metal cations, including Fe(II) and Fe(III) (Benes et al. 1983; von Wirén et al. 1999). The biosynthetic pathway from methionine to NA is conserved in all plant species including graminaceous and non-graminaceous plants (Shojima et al. 1989), where NA may be responsible for metal homeostasis (Hell and Stephan 2003; Takahashi et al. 2003). Consistent with this notion, non-graminaceous plants also possess YS1-like (YSL) genes, some of which are responsible for metal-NA translocation within the plant body (DiDonato et al. 2004; Le Jean et al. 2005; Schaaf et al. 2005; Gendre et al. 2006; Waters et al. 2006; Curie et al. 2008).
Rice (Oryza sativa L.), one of the most important crops for world food supply, takes up Fe from the rhizosphere using the Fe(III)–deoxymugineic acid (DMA) transporter and the ferrous Fe transporter OsIRT1 (Ishimaru et al. 2006). We previously identified 18 putative YS1-like genes (OsYSLs) in the rice genome that exhibited 36–76% sequence similarity to the maize YS1 gene (Koike et al. 2004). We demonstrated that OsYSL2 transports Fe(II)–NA and Mn(II)–NA, but not Fe(III)–DMA (Koike et al. 2004). OsYSL2 expression is strongly induced in Fe-deficient leaves, and is particularly dominant in the phloem cells of leaves and leaf sheaths as well as in developing seeds, suggesting its involvement in phloem transport and the translocation of mineral nutrients in grains (Koike et al. 2004). OsYSL15 transports Fe(III)–DMA, but not Fe(II)–NA, Fe(III)–NA, or Mn(II)–NA (Inoue et al. 2009). OsYSL15 expression is strongly induced in epidermis/exodermis and phloem cells of Fe-deficient roots, suggesting its role in Fe(III)–DMA uptake from the rhizosphere and also in phloem transport of Fe. Analysis of OsYSL15 knockdown plants revealed that OsYSL15 is also crucial in Fe homeostasis during early growth of the seedlings (Inoue et al. 2009). The function of the other OsYSLs has not been investigated, except for the presence of transcripts of OsYSL5, 6, 7, 8, 12, 13, 14, 16, and 17 in roots and OsYSL6, 13, 14, and 16 in leaves (Koike et al. 2004; Inoue et al. 2009). In the present report, we demonstrate that one of the OsYSLs, OsYSL18, encodes a functional Fe(III)–DMA transporter that may be involved in DMA-mediated Fe distribution in the reproductive organs, lamina joints, and phloem cells at the base of the leaf sheath.
Materials and methods
Plant Materials
Non-transgenic and transgenic rice (Oryza sativa cv. Tsukinohikari) seeds were germinated on Murashige and Skoog (MS) medium and transferred to a nutrient solution (Mori and Nishizawa, 1987, Inoue et al. 2003) in a greenhouse with 30°C light/25°C dark periods under natural light conditions. The pH of the culture solution was adjusted daily to 5.5 with 1 M HCl, and the culture solution was renewed weekly. Six-week-old plants were harvested for reverse transcriptase (RT)-PCR and GUS analyses. For Fe, Zn, or Mn deficiency treatments, 4-, 3-, or 2-week-old plants were cultured without each of the specific metals for 2, 3, and 4 weeks, respectively. Flowers and seeds were obtained from soil-grown nutrient-sufficient rice plants.
RT-PCR analysis
Total RNA was isolated from rice plants grown under nutrient-sufficient or nutrient-deficient conditions using an RNeasy Plant Mini Kit (QIAGEN, Tokyo, Japan), and the RNA was treated with RNase-free DNase I (Takara, Tokyo, Japan) to remove contaminating genomic DNA. First-strand cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen, Tokyo, Japan) by priming with oligo-d(T)30. The primers used for RT-PCR were OsYSL18 forward (5′-TAAAGCTGGATGATCCTGAATTCTT) and OsYSL18 reverse (5′-TCGCTCTACATGAAAAGATCAGTTC). These primers were designed to amplify the cDNA across the exons to discriminate possible genomic DNA contamination. The α-tubulin primers used for RT-PCR were α-tubulin forward (5′-TCTTCCACCCTGAGCAGCTC) and α-tubulin reverse (5′-AACCTTGGAGACCAGTGCAG). The sizes of the amplified fragments were confirmed by gel electrophoresis and sequencing. No genomic contamination was observed.
Transport activity of OsYSL18 in Xenopus laevis oocytes
Electrophysiological measurement of OsYSL18 transport activity using Xenopus laevis oocytes was performed according to Igarashi et al. (2000) and Koike et al. (2004). The entire open reading frame (ORF) of OsYSL18 was amplified using two primers: 5′-CACCATGGAGTCGGTCGGCGACCCGCGGG-3′ and 5′- AAGAATTCTAGATCATCCAGCTTTAGCCTTACTCGCCAA-3′. The amplified and verified fragment was subcloned into pENTR/D-TOPO (Invitrogen) to construct pENTR–OsYSL18. The entire ORF of OsYSL18 produced by digestion with EcoRI and XbaI was inserted into the EcoRI and XbaI site of the pGEM-3zf(+) vector. The resulting plasmid, pGEMOsYSL18T, was linearized by XbaI digestion. Capped complementary RNA (cRNA) was synthesized in vitro with a MEGAscript SP6 kit (Ambion, Austin, TX, USA). The oocytes were prepared as described by Igarashi et al. (2000) and injected with 10 ng OsYSL18 cRNA. The injected oocytes were incubated for 2 days and subjected to electrophysiological measurements at pH 7.5. The oocytes were positioned in a 1.0-ml recording chamber perfused with bath solution containing 96 mM NaCl, 2 mM KCl, 5 mM HEPES-NaOH solution (pH 7.5). Membrane currents were measured using the two-microelectrode voltage-clamp method with an automated Hitachi system containing a TEV-200 system (Dagan, Minneapolis, MN, USA). The oocytes were clamped at −80 mV, and steady-state currents in response to the addition of 10 μl metal–chelate complex prepared according to Schaaf et al. (2004) (5 mM final concentration) were obtained. Each oocyte was tested with one substrate. The currents were continuously monitored and analyzed with a Mac Lab system (Adinstruments, Sydney, NSW, Australia). Six independent oocytes injected with OsYSL18 were used to measure the currents. Substrate-induced currents were also measured in six independent water-injected oocytes as controls.
Subcellular localization of OsYSL18-GFP
Using the pH7FWG2 plasmid (Karimi et al. 2002) as the destination vector and pENTR–OsYSL18 as the entry vector, an LR recombination reaction (Invitrogen) between the destination vector and the entry vector generated an expression clone containing the gene encoding cauliflower mosaic virus 35S promoter–OsYSL18–GFP. As a control, a non-tagged green fluorescent protein (GFP) construct CaMV35S–sGFP(S65T)–NOS3′ plasmid (a generous gift from Dr. Yasuo Niwa, University of Shizuoka, Japan) was used. Onion (Allium cepa L.) epidermal cells were transformed using the Biolistic PDS-1000/He Particle Delivery System (Bio-Rad, Tokyo, Japan), and the transiently expressed GFP fluorescence was observed using a laser-scanning confocal microscope (LSM510, Carl Zeiss, Tokyo, Japan) according to Mizuno et al. (2003).
OsYSL18 promoter-GUS analysis
A genomic sequence containing the putative promoter region of OsYSL18 (from −3,000 to −1 bp from the translation initiation codon) was amplified by PCR using genomic DNA as a template. The primers used were: 5′-GAGAGCTAGCGCAGATAACTACTATGCTGCCCTCA-3, and 5′-GAGAGCTAGCCCCGGCCACTTTTCCTCCTCCTCCT-3′. The amplified and verified fragment was excised by HindIII and XbaI and was subcloned upstream of the uidA ORF, which encodes β-glucuronidase (GUS), in the pIG121Hm vector (Hiei et al. 1994). An Agrobacterium tumefaciens strain (C58) carrying the above construct was used to transform rice (Oryza sativa L. cv. Tsukinohikari) as described previously (Higuchi et al. 2001).
T1 and T2 seeds were germinated and cultured as described above and subjected to histochemical GUS analysis following the method of Jefferson et al. (1987) modified by Kosugi et al. (1991). Plant organs and sections were incubated at 37°C for 30 min to overnight in GUS reaction buffer consisting of 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Gluc), 3 mM K4[Fe(CN)6], 0.5 mM K3[Fe(CN)6], 50 mM sodium phosphate buffer (pH 7.0), and 20% (v/v) methanol. After staining, these sections were washed in 70% ethanol for 2 days to remove the chlorophyll and then observed using an Axiophoto microscope (Carl Zeiss) following the manufacturer’s instructions. For in vitro culture of pollen tubes, pollen collected just after anthesis was plated and germinated on medium containing 20% sucrose, 0.32 mM H3BO3, 2.04 mM CaCl2, and 1% agarose. To prepare longitudinal and transverse sections of the basal part of the leaf sheaths, crowns were fixed in 4% formaldehyde, 5% glutaraldehyde, 0.01 M CaCl2, and 0.1 M sodium cacodylate buffer (pH 7.2) for 3 h on ice after staining and chlorophyll removal. Fixed samples were washed three times for 30 min each with 0.1 M sodium cacodylate and 10% sucrose, and then incubated for 2 h on ice in 1% osmium tetroxide and 0.1 M sodium cacodylate. The samples were then serially dehydrated in ethanol, acetone, and propylene oxide, and embedded in Spurr’s resin. Semi-thin sections (5 μm) were cut for observation by light microscopy.
Results
Sequence and structural organization of OsYSL18
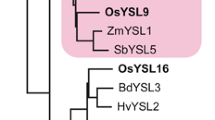
Our search for YS1 homologs in the Oryza sativa L. ssp. japonica (cv. Nipponbare) rice genomic database identified 18 putative OsYSLs that exhibited 36–76% sequence similarity to ZmYS1 (Koike et al. 2004; Fig. 1a). OsYSL18 belongs to a subfamily with no apparent non-graminaceous members, and its cDNA is predicted to have an ORF of 2,040 bp, containing six exons (accession number AB190926). Based on the nucleotide sequence, OsYSL18 encodes a putative protein composed of 679 amino acids (Fig. 1b). OsYSL18 shows 42.5% amino acid identity with the maize YS1 transporter (Curie et al. 2001). We analyzed the topology of OsYSL18 using some web-based programs including SOSUI (http://sosui.proteome.bio.tuat.ac.jp/sosuiframe0.html), which predicted that OsYSL18 has 13 transmembrane domains (TMs) and has its N-terminus localized in the cytoplasm. The abundance of glutamic acid residues at the amino terminus of YS1 (Curie et al. 2001) is not conserved in OsYSL18. The sequence determining substrate specificity of YS1/YSL transporters as reported by Harada et al. (2007) showed low similarity between OsYSL18 and YS1 (Fig. 1b, boxed region).
Sequence characteristics of OsYSL18. a Phylogenetic relationships of YS1-like proteins in rice, maize, barley, and Arabidopsis. Circles represent the four subfamilies. Accession numbers: AB190912 for OsYSL1; AB126253 (AB164646) for OsYSL2; AB190913 for OsYSL3; AB190914 for OsYSL4; AB190915 for OsYSL5; AB190916 for OsYSL6; AB190917 for OsYSL7; AB190918 for OsYSL8; AB190919 for OsYSL9; AB190920 for OsYSL10; AB190921 for OsYSL11; AB190922 for OsYSL12; AB164644 for OsYSL13; AB164645 for OsYSL14; AB190923 for OsYSL15; AB190924 for OsYSL16; AB190925 for OsYSL17; AB190926 for OsYSL18; AF186234 for YS1; AB214183 for HvYS1; At4g24120 for AtYSL1; At5g24380 for AtYSL2; At5g53550 for AtYSL3; At5g41000 for AtYSL4; At3g17650 for AtYSL5; At3g27020 for AtYSL6; At1g65730 for AtYSL7; and At1g48370 for AtYSL8; b comparison of the putative amino acid sequences of OsYSL18 and YS1. White letters in black indicate identical amino acid residues. Lines indicate putative transmembrane domains of OsYSL18 and YS1. The boxed region represents the sequence determining structural properties and substrate specificity of YS1/YSL transporters as reported by Harada et al. (2007)
Transport activity of OsYSL18 in oocytes
We examined the transport activity and substrate specificity of OsYSL18 using Xenopus laevis oocytes. Because previously analyzed YS1 and YSL transporters were shown to be involved in metal homeostasis and transport of DMA- or NA-chelated metals (Curie et al. 2008; Inoue et al. 2009), we investigated whether OsYSL18 was also able to transport DMA- or NA-chelated metals. OsYSL18 was heterologously expressed in the oocytes, and the substrate-induced inward currents at −80 mV in response to Fe(III)–DMA, Fe(II)–NA, Zn(II)–DMA or Zn(II)–NA were measured (Fig. 2). OsYSL18 transported Fe(III)–DMA, but did not transport Fe(II)–NA, Zn(II)–DMA, or Zn(II)–NA.
Transporting activity of OsYSL18 analyzed by electrophysiological measurements using Xenopus laevis oocytes. The transport activities of Fe(III)–DMA, Fe(II)–NA, Zn(II)–DMA, and Zn(II)–NA were measured using the two-electrode voltage-clamp method. The oocytes were clamped at −80 mV, and steady-state currents in response to the addition of a metal–chelate complex (10 μl, 5 mM) were obtained. The data are mean ± SE of six independent oocytes injected with OsYSL18. The same number of water-injected oocytes was used as a control. N.D., not detected
Organ-specific expression pattern of OsYSL18
To investigate the expression pattern of OsYSL18, RT-PCR analysis was conducted using a cDNA pool prepared from the roots, crown, shoots, or flowers of rice plants grown under nutrient-sufficient conditions (Fig. 3). After 45 cycles of PCR, OsYSL18 transcripts were only detected from the cDNA pool prepared from flowers. By increasing the PCR to 55 cycles, OsYSL18 transcripts were also detected in roots, crown, and shoots. No induction of OsYSL18 expression was observed under Fe-, Zn- or Mn-deficient conditions (data not shown).
Spatial patterns of OsYSL18 expression
Subcellular localization of OsYSL18 was analyzed using GFP-fused OsYSL18 protein transiently expressed in onion epidermal cells. Cells expressing the OsYSL18–GFP fusion protein showed fluorescence in the plasma membrane which was clearly distinguishable from the non-fluorescent tonoplast and cytoplasm (Fig. 4a–f). Control experiments demonstrated that cells expressing GFP alone showed signals in the cytoplasm and the nucleus (Fig. 4g–i). Thus, OsYSL18 is a functional transporter for uptake of Fe(III)–DMA in the plasma membrane.
Subcellular localization of OsYSL18 expression. OsYSL18–GFP fusion protein (a–f) or GFP alone (g–i) was transiently expressed in onion epidermal cells and was observed by confocal laser scanning microscopy. (a, d, g) Overlay of confocal cross-sections and transmission images. PM, plasma membrane; T, tonoplast. (b, e, h) Confocal cross-sections. (c, f, i) Transmission images. (d–f) Magnified images of the boxed area shown in (a). Scale bars 20 μm
Cell-type specificity of OsYSL18 expression in whole rice plants was investigated using four independent T1 and T2 lines harboring OsYSL18 promoter (3.0 kb)–GUS constructs, all of which exhibited similar patterns of expression. As OsYSL18 transcripts were most abundant in flowers (Fig. 3), we first examined OsYSL18 promoter activity in flowers and developing seeds (Fig. 5). Before anthesis, the OsYSL18 promoter was active in anthers, pollen, filaments, ovules, stigma, and vascular bundles of the lemma (Fig. 5a). Just after anthesis, expression was also observed in pollen grains and tubes (Fig. 5b, h, i). Expression in germinated pollen was further confirmed using an in vitro culture medium (Fig. 5j). In developing seeds, expression was observed in the embryonic disc and vascular bundles (Fig. 5c–f). In mature seeds, expression was dominant in the embryonic disc (Fig. 5g). OsYSL18 promoter activity was also observed in the embryonic disc of germinating seeds (data not shown).
Cellular localization of OsYSL18 expression in flowers and developing seeds during maturation as observed by histochemical staining of OsYSL18 promoter–GUS expression in transgenic rice plants. Before anthesis (a), just after fertilization (b), and 3 (c), 5 (d), 10 (e), 15 (f), and 30 (g) days after fertilization. (h, i) Magnified image just after fertilization. (j) Germinated pollen tube on in vitro culture medium. Shown are the results from a representative T1 line stained overnight
Because OsYSL18 transcripts were also detected in shoots, crown, and roots, although the expression level was very low in any growth condition (Fig. 3), we then investigated OsYSL18 promoter activity in these organs. Whole-plant GUS staining revealed that OsYSL18 expression was mostly restricted to small regions around lamina joints, crown roots, and the crown (Fig. 6a–e). Around lamina joints, observation of OsYSL18 activity was limited to the basal parts of vascular bundles of leaf blades (Fig. 6a, b). In roots, OsYSL18 was expressed in restricted areas of crown roots (Fig. 6c). Longitudinal sections showed expression in the inner cortex layer above the root tip (Fig. 6d). Around the crown, OsYSL18 activity was observed in the basal region of the leaf sheath (Fig. 6e). Because the crown is a complex position where phloem and xylem are associated, longitudinal and transverse sections around the basal region of leaf sheaths were prepared for further analysis (Fig. 6f–k). OsYSL18 expression was found in the basal regions of vascular bundles in every leaf sheath, but was not found in the shoot apical meristem (Fig. 6f). Expression was mainly observed in phloem cells of phloem parenchyma and companion cells (Fig. 6g–k). These expression patterns were unchanged in Fe-deficient plants (data not shown).
Cellular localization of OsYSL18 expression in vegetative organs as observed by histochemical staining of OsYSL18 promoter–GUS expression in transgenic rice plants. (a) Expression in lamina joints. (b) Magnified image of (a). (c) Expression in crown roots. (d) Longitudinal section of crown roots. (e) Expression in the crown. (f) Longitudinal section around the basal parts of the leaf sheath. Three arrows indicate the positions of the cross-sections shown in (g–j). (g) Cross-section at the upper arrow of (f). The boxed area shows the magnified area in (h). (h–j) Magnified images of cross sections at the three arrows of (f). (h), upper arrow; (i), middle arrow; and (j) lower arrow. Intervals between the images are 600 μm. (k) Magnified image of a cross-section of a vascular bundle. Arrow, phloem companion cell; Xy, xylem. Shown are the results from a representative T1 line stained for 3 h for (a–d), 30 min for (e) and (f), and overnight for (g–k). Scale bars 1 mm for (f–j), and 50 μm for (k)
Discussion
OsYSL18 is a functional Fe(III)–DMA transporter but is not involved in direct Fe uptake from the rhizosphere
To date, YS1, HvYS1, and OsYSL15 have been reported as Fe–MAs transporters in maize, barley, and rice, respectively (Curie et al. 2001; Murata et al. 2006; Inoue et al. 2009). Expression of these transporters is induced by Fe deficiency in roots. Disruption of the YS1 gene leads to leaf chlorosis due to a defect in Fe(III)–MAs uptake (von Wirén et al. 1994). HvYS1 protein is localized to root epidermal cells (Murata et al. 2006). OsYSL15 promoter activity is dominant in epidermal and exodermal cells of Fe-deficient roots (Inoue et al. 2009). According to these data, YS1, HvYS1, and OsYSL15 may be responsible for the primary uptake of Fe–MAs from the rhizosphere. Expression of HvYS1 and OsYSL15 show daily fluctuations (Inoue et al. 2009; Nagasaka et al. 2009), possibly for efficient uptake of Fe coordinated with diurnal secretion of MAs (Takagi et al. 1984). In the present report we have shown that OsYSL18 is another functional Fe(III)–DMA transporter localized to the plasma membrane (Figs. 2, 4). However, the expression pattern of OsYSL18 in roots (Figs. 3, 6) was quite different compared to that of YS1, HvYS1, or OsYSL15. OsYSL18 expression in roots was extremely low and was not upregulated by Fe deficiency. OsYSL18 promoter activity was not observed in whole roots but was specific to crown roots (Fig. 6c). Moreover, OsYSL18 promoter activity in crown roots was restricted to the inner cortex layer (Fig. 6d), but was not obvious in epidermal and exodermal cells, where absorption of Fe(III)–DMA mainly occurs. These results suggest that OsYSL18 is a functional Fe(III)–DMA transporter, but is not involved in primary uptake of Fe(III)–DMA from the rhizosphere. Thus, the rice Strategy-II Fe uptake system would be primarily attributed to OsYSL15 but not to OsYSL18. The primary function of OsYSL18 is deduced to be translocation of Fe(III)–DMA within the plant body.
Role of OsYSL18 in fertilization
Some members of the YSL family are reported to be involved in metal movement to floral tissues mediated by NA (Curie et al. 2008). In addition to NA, DMA also exists in developing and mature rice grains (Takahashi et al. 2006; unpublished). Interestingly, knockdown mutants of OsYSL15 exhibit severe arrest in germination and early growth, which are rescued by high iron supply (Inoue et al. 2009), suggesting that Fe in rice endosperm is stored at least partly in the form of Fe(III)–DMA to be utilized via OsYSL15 during seed germination. OsYSL18 transcripts were most abundant in flowers (Fig. 3) and OsYSL18 promoter activity was observed in reproductive organs before and after anthesis (Fig. 5). These results strongly suggest the involvement of OsYSL18 in Fe transport to reproductive organs. Along with OsYSL18, OsYSL2 and OsYSL15 are co-expressed in vascular bundles of the lemma and embryonic disc (Koike et al. 2004; Inoue et al. 2009). In these organs, these Fe(II)–NA and Fe(III)–DMA transporters may be cooperating in Fe translocation into seeds. Remarkably, the OsYSL18 promoter is also active in pollen tubes (Fig. 5b, h–j). Our previous examination of other transporters involved in Fe homeostasis clarified that OsFRDL1, a citrate effluxer required for efficient Fe translocation (Yokosho et al. 2009), is also expressed in pollen grains (Inoue et al. 2004), whereas none of the other known Fe-related transporter genes including OsYSL2, OsYSL15, and ferrous transporter gene OsIRT1 were expressed in pollen tubes (Koike et al. 2004; Inoue et al. 2009; Takahashi et al. unpublished). In Arabidopsis, two YSL genes, AtYSL1 and AtYSL3, are expressed in pollen grains, possibly mediating metal-NA translocation (Le Jean et al. 2005; Waters et al. 2006). Elongation of pollen tubes is indispensable for fertilization, during which Fe is required for various vital processes, including respiration, as well as many other enzymatic reactions. Our results strongly suggest that OsYSL18 plays an important role in pollen function and fertilization by transporting Fe(III)–DMA into pollen cells.
OsYSL18 functions in the crown and lamina joints
In vegetative shoot organs, OsYSL18 expression was specifically observed around the crown and lamina joints (Fig. 6a, b, e). In the crown, OsYSL18 was expressed only in the basal parts of leaf sheath vascular bundles (Fig. 6f). Furthermore, careful observation of transverse sections revealed that OsYSL18 expression was restricted to the phloem parenchyma and companion cells (Fig. 6g–k). This suggests that OsYSL18 is involved in the loading of Fe(III)–DMA to phloem by transporting the metal complexes to phloem parenchyma and companion cells. Interestingly, previous tracer experiments showed that various elements supplied from roots or leaves of barley and rice first accumulate in the basal parts of the shoot and are then distributed to other plant parts, as determined in studies using 59Fe (Mori, 1998), 52Fe, 52Mn, and 62Zn (Watanabe et al. 2001; Tsukamoto et al. 2006, 2009), 11 C-methionine (Nakanishi et al. 1999; Bughio et al. 2001), 13N (Kiyomiya et al. 2001a), and H 152 O (Mori et al. 2000; Kiyomiya et al. 2001b; Nakanishi et al. 2002; Tsukamoto et al. 2004). As a result of these studies, the basal part of the shoot was designated as a discrimination center (DC), where phloem and xylem structures are complexly associated, and minerals and metabolites accumulate prior to translocation to other plant parts. Recently, Tsukamoto et al. (2009) reported that 52Fe supplied as 52Fe(III)–DMA to roots of barley is translocated mainly via phloem to the youngest leaf, whereas it is translocated mostly via xylem to older leaves, suggesting the importance of Fe transfer from xylem to phloem in the DC and/or the roots. OsYSL18 expression was specific to the region very close to DC. Therefore, phloem loading of Fe(III)–DMA via OsYSL18 might be important for Fe distribution to leaf sheaths, especially where xylem Fe translocation is not fully developed.
In lamina joints, the OsYSL18 expression pattern was very similar to that of the crown. Expression was observed only in vascular bundles at the base of leaf blades (Fig. 6a, b). To date, there have been no reports regarding any special role of rice Fe nutrition in lamina joints. The expression of the Fe(III)–DMA transporter specific to the basal parts of vascular bundles of the leaf blades indicates that the lamina joint might also play an important role in Fe translocation in rice. Recently, Tanaka et al. (2008) reported that an Arabidopsis boric acid channel NIP6;1 is predominantly expressed in phloem cells of nodal regions, where xylem–phloem transfer of boric acid is proposed to occur. It is conceivable that vascular plants have developed various joint structures to facilitate xylem–phloem transfer of nutritional elements.
Involvement of DMA and NA in the long-distance transport of Fe
Among various metal chelators, the prominent importance of NA and citrate in phloem and xylem Fe translocation have been revealed by physiological, genetic, and molecular studies both in graminaceous and non-graminaceous species (Hell and Stephan 2003; Takahashi et al. 2003; Durrett et al. 2007; Curie et al. 2008; Yokosho et al. 2009). In contrast to these Fe chelators, MAs historically have been identified and characterized as substances responsible for Fe acquisition from the rhizosphere rather than internal Fe translocation. Nevertheless, various lines of evidence suggest that MAs also play roles for internal Fe distribution in graminaceous plants. Endogenous MAs were detected in shoots of barley and rice, and the amount of MAs increased dramatically under Fe deficiency (Higuchi et al. 2001). DMA was also detected in rice phloem sap (Mori et al. 1991). The genes involved in DMA biosynthesis in rice, OsNAS1-3, OsNAAT1, and OsDMAS1, are co-expressed in the phloem companion cells in roots and leaves (Inoue et al. 2003, 2008; Bashir et al. 2006). Furthermore, the OsYSL15 transporter gene is also expressed in the phloem companion cells of roots and leaves, as well as in reproductive organs (Inoue et al. 2009). Zn might also be internally transported as a MAs-chelated form, because Zn(II)–DMA is translocated to Zn-deficient rice leaves at a higher rate than Zn2+ (Suzuki et al. 2008). Expression of the newly found Fe(III)–DMA transporter gene, OsYSL18, in the phloem cells of phloem parenchyma and companion cells (Fig. 6) strongly supports the notion of the involvement of DMA in long-distance transport of Fe via phloem. Although Fe(II)–NA complexes are predicted to be more stable than Fe(III)–DMA in terms of phloem sap pH (von Wirén et al. 1999), and thus chelation is likely to change from DMA to NA in the phloem cells, MAs are likely to play roles for phloem Fe translocation as well. OsYSL2 and OsYSL15, as well as OsYSL18, are also co-expressed in large areas of flowers and developing seeds (Fig. 5; Koike et al. 2004; Inoue et al. 2009). Thus, both Fe(II)–NA and Fe(III)–DMA also may be involved in seed Fe loading.
YSL genes consist of a large family, in which the physiological role of each member would be dependent both on its expression pattern and substrate specificity. In rice, expression of OsYSL2 and OsYSL15 in vegetative organs is strongly upregulated under Fe deficiency (Koike et al. 2004; Inoue et al. 2009), whereas OsYSL18 is expressed constitutively, with no induction in response to Fe deficiency. This evidence suggests that OsYSL2 and OsYSL15 play important roles in Fe utilization, especially under low Fe availability, whereas OsYSL18 is responsible for basic Fe homeostasis, which might not be fully accomplished by OsYSL2 and OsYSL15 under normal nutritional conditions. Expression of OsYSL18 in specific cells, as discussed above, suggests the fundamental role of the Fe(III)–DMA transport in these cells, including pollen and phloem cells at the base of the leaf sheath.
Maize YS1 transporter is able to transport not only Fe(III)–DMA, but also many other DMA- and NA-chelated metals, including Fe(II)–NA (Schaaf et al. 2004). Fe-deficiency-induced expression of YS1 is evident both in roots and shoots (Curie et al. 2001; Roberts et al. 2004), suggesting that YS1 is responsible not only for Strategy-II Fe uptake, but also for internal transport of Fe as either MAs- or NA-chelated forms. It might be interesting to explore why rice utilizes at least three YSL members (OsYSL2, OsYSL15, and OsYSL18) to accomplish Fe utilization, which might be mediated by a single YS1 transporter in maize.
YSL family is phylogenetically composed of four subfamilies. OsYSL18 belongs to the only subfamily with no apparent non-graminaceous members (Fig. 1a; Curie et al. 2008). This evidence, along with the result that OsYSL18 transports Fe(III)–DMA but not Fe(II)–NA (Fig. 2), suggests the possibility that this subfamily of YSL transporters have been evolved to perform graminaceous-specific functions, which might be accomplished by transporting Fe(III)–MAs specifically but not Fe(II)–NA. Harada et al. (2007) reported that the outer membrane loop between the sixth and seventh transmembrane regions of YS1 and HvYS1 is essential for substrate specificity. They found 20 amino acid residues that maintain the helical propensity of HvYS1, which is deduced to be responsible for the more extreme substrate specificity of HvYS1 compared to YS1. Among these 20 amino acid residues, OsYSL18 possesses a sequence with little similarity to YS1 (4/20 residues; Fig. 1b, boxed region), whereas it has more similarity to HvYS1 (8/20 residues) or OsYSL15 (7/20 residues). This evidence favors strict substrate specificity of OsYSL18 (Fig. 2), which is more similar to that of HvYS1 and OsYSL15 than YS1. Because the OsYSL18 sequence shares relatively low similarity to any other YS1/YSL transporters in which substrate specificity has been determined, OsYSL18 and other members of this subfamily of YSL might form structures distinct from other well-known YS1/YSL members. Further analysis of transport activity, in combination with domain swap analysis, mutagenesis, or X-ray crystal structural analysis, of these YS1/YSL transporters should reveal the molecular mechanisms underlying substrate recognition and specificity.
The YSL family is also distantly related to the large group of oligopeptide transporters (OPT), among which an Arabidopsis gene (AtOPT3) is upregulated under Fe deficiency and is involved in Fe translocation to developing seeds (Stacey et al. 2006, 2008). Functional characterization, including substrate specificity, of AtOPT3 and its counterparts in graminaceous species, would be beneficial for better understanding of Fe translocation and utilization in plants. Finally, manipulation of Fe(III)–DMA transporters in combination with other key components controlling Fe homeostasis will provide further understanding of the mechanisms of Fe nutrition and production of Fe-fortified rice crops.
References
Bashir K, Inoue H, Nagasaka S, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2006) Cloning and characterization of deoxymugineic acid synthase genes from graminaceous plants. J Biol Chem 281:32395–32402. doi:10.1074/jbc.M604133200
Benes I, Schreiber K, Ripperger H, Kircheiss A (1983) Metal complex formation by nicotianamine, a possible phytosiderophore. Experientia 39:261–262. doi:10.1007/BF01955293
Bughio N, Nakanishi H, Kiyomiya S, Matsuhashi S, Ishioka N, Watanabe S, Uchida H, Tsuji A, Osa A, Kume T, Hashimoto S, Sekine T, Mori S (2001) Real-time [11C]methionine translocation in barley in relation to mugineic acid family phytosiderophores. Planta 213:708–715. doi:10.1007/s004250100552
Curie C, Panavience Z, Loulergue C, Dellaporta SL, Briat JF, Walker EL (2001) Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409:346–349. doi:10.1038/35053080
Curie C, Cassin G, Couch D, Divol F, Higuchi K, Le Jean M, Misson J, Schikora A, Czernic P, Mari S (2008) Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Ann Bot (Lond) 103:1–11. doi:10.1093/aob/mcn207
DiDonato RJ Jr, Roberts LA, Sanderson T, Eisley RB, Walker EL (2004) Arabidopsis Yellow Stripe-Like2 (YSL2): a metal-regulated gene encoding a plasma membrane transporter of nicotianamine-metal complexes. Plant J 39:403–414. doi:10.1111/j.1365-313X.2004.02128.x
Durrett TP, Gassmann W, Rogers EE (2007) The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol 144:197–205. doi:10.1104/pp.107.097162
Eide D, Broderuis M, Fett J, Guerinot ML (1996) A novel iron regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93:5624–5628. doi:10.1073/pnas.93.11.5624
Gendre D, Czernic P, Conéjéro G, Pianelli K, Briat JF, Lebrun M, Mari S (2006) TcYSL3, a member of the YSL gene family from the hyperaccumulator Thlaspi caerulescens, encodes a nicotianamine-Ni/Fe transporter. Plant J 49:1–15. doi:10.1111/j.1365-313X.2006.02937.x
Grusak MA, Dellapenna D (1999) Improving the nutrient composition of plants to enhance human nutrition and health. Annu Rev Plant Physiol Plant Mol Biol 50:133–161. doi:10.1146/annurev.arplant.50.1.133
Harada E, Sugase K, Namba K, Iwashita T, Murata Y (2007) Structural element responsible for the Fe(III)-phytosiderophore specific transport by HvYS1 transporter in barley. FEBS Lett 581:4298–4302. doi:10.1016/j.febslet.2007.08.011
Hell R, Stephan UW (2003) Iron uptake, trafficking and homeostasis in plants. Planta 216:541–551
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282. doi:10.1046/j.1365-313X.1994.6020271.x
Higuchi K, Watanabe S, Takahashi M, Kawasaki S, Nakanishi H, Nishizawa NK, Mori S (2001) Nicotianamine synthase gene expression differs in barley and rice under Fe-deficient conditions. Plant J 25:159–167. doi:10.1046/j.1365-313x.2001.00951.x
Igarashi Y, Yoshiba Y, Takeshita T, Nomura S, Otomo J, Yamaguchi-Shinozaki K, Shinozaki K (2000) Molecular cloning and characterization of a cDNA encoding proline transporter in rice. Plant Cell Physiol 41:750–756. doi:10.1093/pcp/pcd015
Inoue H, Higuchi K, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2003) Three rice nicotianamine synthase genes, OsNAS1, OsNAS2 and OsNAS3 are expressed in cells involved in long-distance transport of iron and differentially regulated by iron. Plant J 36:366–381. doi:10.1046/j.1365-313X.2003.01878.x
Inoue H, Suzuki M, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2004) A rice FRD3-like (OsFRDL1) gene is expressed in the cells involved in long-distance transport. Soil Sci Plant Nutr 50:1133–1140
Inoue H, Takahashi M, Kobayashi T, Suzuki M, Nakanishi H, Mori S, Nishizawa NK (2008) Identification and localisation of the rice nicotianamine aminotransferase gene OsNAAT1 expression suggests the site of phytosiderophore synthesis in rice. Plant Mol Biol 66:193–203. doi:10.1007/s11103-007-9262-8
Inoue H, Kobayashi T, Nozoye T, Takahashi M, Kakei Y, Suzuki K, Nakazono M, Nakanishi H, Mori S, Nishizawa NK (2009) Rice OsYSL15 is an iron-regulated iron(III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J Biol Chem 284:3470–3479. doi:10.1074/jbc.M806042200
Ishimaru Y, Suzuki M, Tsukamoto T, Suzuki K, Nakazono M, Kobayashi T, Wada Y, Watanabe S, Matsuhashi S, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2006) Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J 45:335–346. doi:10.1111/j.1365-313X.2005.02624.x
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Karimi M, Inze D, Depicker A (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7:193–195. doi:10.1016/S1360-1385(02)02251-3
Kiyomiya S, Nakanishi H, Uchida H, Tsuji A, Nishiyama S, Futatsubashi M, Tsukada H, Ishioka NS, Watanabe S, Ito T, Mizuniwa C, Osa A, Matsuhashi S, Hashimoto S, Sekine T, Mori S (2001a) Real time visualization of 13 N-translocation in rice under different environmental conditions using a positron-emitting tracer imaging system. Plant Physiol 125:1743–1754. doi:10.1104/pp.125.4.1743
Kiyomiya S, Nakanishi H, Uchida H, Nishiyama S, Tsukada H, Ishioka NS, Watanabe S, Osa A, Mizuniwa C, Ito T, Matsuhashi S, Hashimoto S, Sekine T, Tsuji A, Mori S (2001b) Light activates H 152 O flow in rice: detailed monitoring using a positron-emitting tracer imaging system (PETIS). Physiol Plant 113:359–367. doi:10.1034/j.1399-3054.2001.1130309.x
Koike S, Inoue H, Mizuno D, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2004) OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J 39:415–424. doi:10.1111/j.1365-313X.2004.02146.x
Kosugi S, Suzuka I, Ohashi Y, Murakami T, Arai Y (1991) Upstream sequences of rice proliferating cell nuclear antigen (PCNA) gene mediate expression of PCNA-GUS chimeric gene in meristems of transgenic tobacco plants. Nucleic Acids Res 19:1571–1576. doi:10.1093/nar/19.7.1571
Le Jean M, Schikora A, Mari S, Briat JF, Curie C (2005) A loss-of-function mutation in AtYSL1 reveals its role in iron and nicotianamine seed loading. Plant J 44:769–782. doi:10.1111/j.1365-313X.2005.02569.x
Mason JB, Lotfi M, Dalmiya N, Sethuraman K, Deitchler M, Geibel S, Gillenwater K, Gilman A, Mason K, Mock N (2001) The micronutrient report: Current progress and trends in the control of vitamin A, iodine, and iron deficiencies. The Micronutrient Initiative/International Development Research Centre, Ottawa, Canada
Mizuno D, Higuchi K, Sakamoto T, Nakanishi H, Mori S, Nishizawa NK (2003) Three nicotianamine synthase genes isolated from maize are differentially regulated by iron nutritional status. Plant Physiol 132:1989–1997. doi:10.1104/pp.102.019869
Mori S (1998) Iron transport in graminaceous plants. In: Sigel A, Sigel H (eds) Iron transport and storage in microorganisms, plants and animals, vol 35. Marcel Dekker, New York, pp 215–237
Mori S (1999) Iron acquisition by plants. Curr Opin Plant Biol 2:250–253. doi:10.1016/S1369-5266(99)80043-0
Mori S, Nishizawa N (1987) Methionine as a dominant precursor of phytosiderophores in graminaceae plants. Plant Cell Physiol 28:1081–1092
Mori S, Nishizawa N, Hayashi H, Chino M, Yoshimura E, Ishihara J (1991) Why are young rice plants highly susceptible to iron deficiency? Plant Soil 130:143–156. doi:10.1007/BF00011869
Mori S, Kiyomiya S, Nakanishi H, Ishioka N, Watanabe S, Osa A, Matsuhashi S, Hashimoto S, Sekine T, Uchida H, Nishiyama S, Tsukada H, Tsuji A (2000) Visualization of 15O-water flow in tomato and rice in the light and dark using a positron-emitting tracer imaging system (PETIS). Soil Sci Plant Nutr 46:975–979
Murata Y, Ma JF, Yamaji N, Ueno D, Nomoto K, Iwashita T (2006) A specific transporter for iron(III)-phytosiderophore in barley roots. Plant J 46:563–572. doi:10.1111/j.1365-313X.2006.02714.x
Nagasaka S, Takahashi M, Itai RN, Bashir K, Nakanishi H, Mori S, Nishizawa NK (2009) Time course analysis of gene expression over 24 h in Fe-deficient barley roots. Plant Mol Biol 69:621–631. doi:10.1007/s11103-008-9443-0
Nakanishi H, Bughio N, Matsuhashi S, Ishioka N, Uchida H, Tsuji A, Osa A, Sekine T, Kume T, Mori S (1999) Visualizing real time [11C]methionine translocation in Fe-sufficient and Fe-deficient barley using a positron emitting tracer imaging system (PETIS). J Exp Bot 50:637–643. doi:10.1093/jexbot/50.334.637
Nakanishi H, Kiyomiya S, Tsukamoto T, Tsukada H, Uchida H, Mori S (2002) Water (H 152 O) flow in rice is regulated by the concentration of nutrient as monitored by positron multi-probe system (PMPS). Soil Sci Plant Nutr 48:759–762
Roberts LA, Pierson AJ, Panaviene Z, Walker EL (2004) Yellow stripe1. Expanded roles for the maize iron-phytosiderophore transporter. Plant Physiol 135:112–120. doi:10.1104/pp.103.037572
Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397:694–697. doi:10.1038/17800
Römheld V, Marschner H (1986) Evidence for a specific uptake system for iron phytosiderophores in roots of grasses. Plant Physiol 80:175–180. doi:10.1104/pp.80.1.175
Schaaf G, Ludewig U, Erenoglu BE, Mori S, Kitahara T, von Wirén N (2004) ZmYS1 functions as a proton-coupled symporter for phytosiderophore- and nicotianamine-chelated metals. J Biol Chem 279:9091–9096. doi:10.1074/jbc.M311799200
Schaaf G, Schikora A, Haberle J, Vert G, Ludewig U, Briat JF, Curie C, von Wirén N (2005) A putative function for the Arabidopsis Fe-phytosiderophore transporter homolog AtYSL2 in Fe and Zn homeostasis. Plant Cell Physiol 46:762–774. doi:10.1093/pcp/pci081
Shojima S, Nishizawa NK, Mori S (1989) Establishment of a cell-free system for the biosynthesis of nicotianamine. Plant Cell Physiol 30:673–677
Shojima S, Nishizawa NK, Fushiya S, Nozoe S, Irifune T, Mori S (1990) Biosynthesis of phytosiderophores. In vitro biosynthesis of 2′-deoxymugineic acid from L-methionine and nicotianamine. Plant Physiol 93:1497–1503. doi:10.1104/pp.93.4.1497
Stacey MG, Osawa H, Patel A, Gassmann W, Stacey G (2006) Expression analysis of Arabidopsis oligopeptide transporter during seed germination, vegetative growth and reproduction. Planta 223:291–305. doi:10.1007/s00425-005-0087-x
Stacey MG, Patel A, McClain WE, Mathieu M, Remley M, Rogers EE, Gassmann W, Blevins DG, Stacey G (2008) The Arabidopsis OPT3 protein functions in metal homeostasis and movement of iron to developing seeds. Plant Physiol 146:589–601. doi:10.1104/pp.107.108183
Suzuki M, Tsukamoto T, Inoue H, Watanabe S, Matsuhashi S, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2008) Deoxymugineic acid increases Zn translocation in Zn-deficient rice plants. Plant Mol Biol 66:609–617. doi:10.1007/s11103-008-9292-x
Takagi S (1976) Naturally occurring iron-chelating compounds in oat- and rice-root washings. Soil Sci Plant Nutr 22:423–433
Takagi S, Nomoto K, Takemoto T (1984) Physiological aspect of mugineic acid, a possible phytosiderophore of graminaceous plants. J Plant Nutr 7:469–477. doi:10.1080/01904168409363213
Takahashi M, Terada Y, Nakai I, Nakanishi H, Yoshimura E, Mori S, Nishizawa NK (2003) Role of nicotianamine in the intracellular delivery of metals and plant reproductive development. Plant Cell 15:1263–1280. doi:10.1105/tpc.010256
Takahashi M, Inoue H, Ishimaru Y, Nakanishi H, Mori S, Nishizawa NK (2006) The role of nicotianamine and mugineic acid in metal transport at reproductive stage. Plant Cell Physiol 47:s230
Tanaka M, Wallace IS, Takano J, Roberts DM, Fujiwara T (2008) NIP6;1 is a boric acid channel for preferential transport of boron to growing shoot tissues in Arabidopsis. Plant Cell 20:2860–2875. doi:10.1105/tpc.108.058628
Tsukamoto T, Uchida H, Nakanishi H, Nishiyama S, Tsukada H, Matsuhashi S, Nishizawa NK, Mori S (2004) H 152 O translocation in rice was enhanced by 10 μM 5-aminolevulinic acid as monitored by positron emitting tracer imaging system (PETIS). Soil Sci Plant Nutr 50:1085–1088
Tsukamoto T, Nakanishi H, Kiyomiya S, Watanabe S, Matsuhashi S, Nishizawa NK, Mori S (2006) 52Mn translocation in barley monitored using a positron-emitting tracer imaging system. Soil Sci Plant Nutr 52:717–725. doi:10.1111/j.1747-0765.2006.00096.x
Tsukamoto T, Nakanishi H, Uchida H, Watanabe S, Matsuhashi S, Mori S, Nishizawa NK (2009) 52Fe translocation in barley as monitored by a positron emitting tracer imaging system (PETIS): evidence for the direct translocation of Fe from roots to young leaves via phloem. Plant Cell Physiol 50:48–57. doi:10.1093/pcp/pcn192
von Wirén N, Mori S, Marschner H, Römheld V (1994) Iron inefficiency in maize mutant ys1 (Zea mays L cv yellow-stripe) is caused by a defect in uptake of iron phytosiderophores. Plant Physiol 106:71–77
von Wirén N, Klair S, Bansal S, Briat JF, Khodr H, Shioiri T, Leigh RA, Hider RC (1999) Nicotianamine chelates both FeIII and FeII Implications for metal transport in plants. Plant Physiol 119:1107–1114. doi:10.1104/pp.119.3.1107
Watanabe S, Ishioka NS, Osa A, Koizumi M, Sekine T, Kiyomiya S, Nakanishi H, Mori S (2001) Production of positron emitters of metallic elements to study plant uptake and distribution. Radiochim Acta 89:853–858. doi:10.1524/ract.2001.89.11-12.853
Waters BM, Chu HS, DiDonato RJ Jr, Roberts LA, Eisley RB, Lahner B, Salt D, Walker EL (2006) Mutations in Arabidopsis Yellow Stripe-Like1 (YSL1) and Yellow Stripe-Like3 (YSL3) reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiol 141:1446–1458. doi:10.1104/pp.06.082586
Yokosho K, Yamaji N, Ueno D, Mitani N, Ma JF (2009) OsFRDL1 is a citrate transporter required for efficient translocation of iron in rice. Plant Physiol 149:297–305. doi:10.1104/pp.108.128132
Acknowledgments
We are indebted to Dr. Yoshiaki Nagamura (the Rice Genome Project and the NIAS DNA Bank) for providing the rice cDNA clone, Dr. Yasuo Niwa (University of Shizuoka) for providing the CaMV35S–sGFP(S65T)–NOS3’ plasmid, Dr. Takeshi Kitahara (The University of Tokyo) for providing chemically synthesized NA and DMA, and Dr. Khurram Bashir (The University of Tokyo) for carefully reading the manuscript. This research was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and Core Research for Evolutional Science and Technology (CREST), Japan Science and Technology Agency (JST).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Accession numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession number AB190926 for OsYSL18.
Takahiro Aoyama and Takanori Kobayashi contributed equally to this work.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Aoyama, T., Kobayashi, T., Takahashi, M. et al. OsYSL18 is a rice iron(III)–deoxymugineic acid transporter specifically expressed in reproductive organs and phloem of lamina joints. Plant Mol Biol 70, 681–692 (2009). https://doi.org/10.1007/s11103-009-9500-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-009-9500-3