Abstract
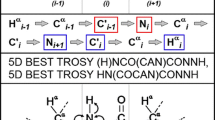
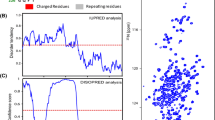
A pair of 4D NMR experiments for the backbone assignment of disordered proteins is presented. The experiments exploit 13C direct detection and non-uniform sampling of the indirectly detected dimensions, and provide correlations of the aliphatic proton (Hα, and Hβ) and carbon (Cα, Cβ) resonance frequencies to the protein backbone. Thus, all the chemical shifts regularly used to map the transient secondary structure motifs in the intrinsically disordered proteins (Hα, Cα, Cβ, C′, and N) can be extracted from each spectrum. Compared to the commonly used assignment strategy based on matching the Cα and Cβ chemical shifts, inclusion of the Hα and Hβ provides up to three extra resonance frequencies that decrease the chance of ambiguous assignment. The experiments were successfully applied to the original assignment of a 12.8 kDa intrinsically disordered protein having a high content of proline residues (26 %) in the sequence.







Similar content being viewed by others
References
Atreya H, Eletsky A, Szyperski T (2005) Resonance assignment of proteins with high shift degeneracy based on 5D spectral information encoded in G(2)FT NMR experiments. J Am Chem Soc 127(13):4554–4555
Barna JCJ, Laue ED, Mayger MR, Skilling J, Worrall SJP (1987) Exponential sampling: an alternative method for sampling in two-dimensional NMR experiments. J Magn Reson 73(1):69–77
Barna JCJ, Laue ED (1987) Comparison of conventional and exponential sampling for 2D NMR experiments: application to a 2D NMR spectrum of a protein. J Magn Reson 75:384–389
Bermel W, Bertini I, Duma L, Felli IC, Emsley L, Pierattelli R, Vasos PR (2005) Complete assignment of heteronuclear protein resonances by protonless NMR spectroscopy. Angew Chem Int Ed 44(20):3089–3092
Bermel W, Bertini I, Felli IC, Piccioli M, Pierattelli R (2006) C-13-detected protonless NMR spectroscopy of proteins in solution. Prog Nucl Mag Res Sp 48(1):25–45
Bermel W, Bertini I, Felli I, Lee Y, Luchinat C, Pierattelli R (2006) Protonless NMR experiments for sequence-specific assignment of backbone nuclei in unfolded proteins. J Am Chem Soc 128(12):3918–3919
Bermel W, Bertini I, Felli IC, Kümmerle R, Pierattelli R (2006) Novel 13C direct detection experiments, including extension to the third dimension, to perform the complete assignment of proteins. J Magn Reson 178(1):56–64
Bermel W, Bertini I, Csizmok V, Felli IC, Pierattelli R, Tompa P (2009) H-start for exclusively heteronuclear NMR spectroscopy: the case of intrinsically disordered proteins. J Magn Reson 198(2):275–281
Bermel W, Bertini I, Felli IC, Piccioli M, Pierattelli R (2009) Speeding up C-13 direct detection biomolecular NMR spectroscopy. J Am Chem Soc 131(42):15339–15345
Billadeau DD, Burkhardt JK (2006) Regulation of cytoskeletal dynamics at the immune synapse: new stars join the actin troupe. Traffic 7(11):1451–1460
Bodenhausen G, Ernst RR (1981) The accordion experiment, a simple approach to three dimensional NMR spectroscopy. J Magn Reson 45:367–373
Bodenhausen G, Ernst RR (1982) Direct determination of rate constants of slow dynamic processes by two-dimensional “accordion” spectroscopy in nuclear magnetic resonance. J Am Chem Soc 104(5):1304–1309
Brutscher B (2002) Intraresidue HNCA and COHNCA experiments for protein backbone resonance assignment. J Magn Reson 156(1):155–159
Bussell R, Eliezer D (2001) Residual structure and dynamics in Parkinson’s disease-associated mutants of alpha-synuclein. J Biol Chem 276(49):45996–46003
Cai M, Ying H, Sakaguchi K, Clore GM, Gronenborn AM, Craigie R (1998) An efficient and cost eddective isotope labeling protocol for proteins expressed in shape Escherichia coli. J Biomol NMR 11(1):97–102
Coggins BE, Zhou P (2007) Sampling of the time domain along concentric rings. J Magn Reson 184:207–221
Coggins BE, Zhou P (2008) High resolution 4-D spectroscopy with sparse concentric shell sampling and FFT-CLEAN. J Biomol NMR 42(4):225–239
de la Fuente MA, Sasahara Y, Calamito M, Anton IM, Elkhal A, Gallego MD, Suresh K, Siminovitch K, Ochs HD, Anderson KC, Rosen FS, Geha RS, Ramesh N (2007) WIP is a chaperone for Wiskott-Aldrich syndrome protein (WASp). PNAS 104(3):926–931
Ding K, Gronenborn AM (2008) Novel 2D triple-resonance NMR experiments for sequential resonance assignments of proteins. J Magn Reson 156:262–268
Delaglio F, Grzesiek S, Vuister G, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6(3):277–293
Derry JM, Ochs HD, Francke U (1994) Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell 78(4):635–644
Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ (2000) Intrinsic protein disorder in complete genomes. Genome Inform 11:161–171
Dunker AK, Silman I, Uversky VN, Sussman JL (2008) Function and structure of inherently disordered proteins. Curr Opin Struct Biol 18(6):756–764
Dyson H, Wright P (2004) Unfolded proteins and protein folding studied by NMR. Chem Rev 104(8):3607–3622
Dyson H, Wright P (2005) Intrinsically unstructured proteins and their functions. Nature Rev Mol Cell Biol 6(3):197–208
Eliezer D (2007) Characterizing residual structure in disordered protein states using nuclear magnetic resonance. Methods Mol Biol 350:49–67
Eliezer D (2009) Biophysical characterization of intrinsically disordered proteins. Curr Opin Struct Biol 19(1):23–30
Fink A (2005) Natively unfolded proteins. Curr Opin Struct Biol 15(1):35–41
Frueh DP, Sun ZYJ, Vosburg DA, Walsh CT, Hoch JC, Wagner G (2006) Non-uniformly sampled double-TROSY hNcaNH experiments for NMR sequential assignments of large proteins. J Am Chem Soc 128(17):5757–5763
Ganguly D, Chen J (2009) Structural interpretation of paramagnetic relaxation enhancement-derived distances for disordered protein states. J Mol Biol 390(3):467–477
Hiller S, Fiorito F, Wuthrich K, Wider G (2005) Automated projection spectroscopy (APSY). PNAS 102(31):10876–10881
Hiller S, Wasmer C, Wider G, Wüthrich K (2007) Sequence-specific resonance assignment of soluble nonglobular proteins by 7D APSY-NMR spectroscopy. J Am Chem Soc 129(35):10,823–10,828
Kazimierczuk K, Koźmiński W, Zhukov I (2006) Two-dimensional Fourier transform of arbitrarily sampled NMR data sets. J Magn Reson 179:323–328
Kazimierczuk K, Zawadzka A, Koźmiński W (2008) Optimization of random time domain sampling in multidimensional NMR. J Magn Reson 192(1):123–130
Kazimierczuk K, Zawadzka A, Koźmiński W (2009) Narrow peaks and high dimensionalities: exploiting the advantages of random sampling. J Magn Reson 205(2):286–292
Kazimierczuk K, Zawadzka-Kazimierczuk A, Koźmiński W (2010) Non-uniform frequency domain for optimal exploitation of non-uniform sampling. J Magn Reson 197(2):219–228
Kim S, Szyperski T (2003) GFT NMR, a new approach to rapidly obtain precise high-dimensional NMR spectra information. J Am Chem Soc 125:1385–1393
Kupče Ē, Freeman R (2003) Projection-reconstruction of three-dimensional NMR spectra. J Am Chem Soc 125:13958–13959
Kupče Ē, Freeman R (2008) Hyperdimensional NMR spectroscopy. Prog Nucl Magn Reson 11:22–30
Malmodin D, Billeter M (2005) Multiway decomposition of NMR spectra with coupled evolution periods. J Am Chem Soc 127(39):13486–13487
Marion D (2006) Processing of ND NMR spectra sampled in polar coordinates: a simple Fourier transform instead of a reconstruction. J Biomol NMR 36(1):45–54
Mobli M, Stern AS, Hoch JC (2008) Spectral reconstruction methods in fast NMR: reduced dimensionality, random sampling and maximum entropy. J Magn Reson 182(1):96–105
Mobli M, Hoch JC (2008) Maximum entropy spectral reconstruction of nonuniformly sampled data. Concepts Magn Reson 32(6):436–448
Motáčková V, Kubíčková M, Kožíšek M, Grantz-Šašková K, Švec M, Žídek L, Sklenář V (2009) Backbone H-1, C-13, and N-15 NMR assignment for the inactive form of the retroviral protease of the murine intracisternal A-type particle, inMIA-14 PR. Biomol NMR Assignments 3(2):261–264
Motáčková V, Nováček J, Zawadzka-Kazimierczuk A, Kazimierczuk K, Žídek L, Koźmiński W, Sklenář V (2010) Strategy for complete NMR assignment of disordered proteins with highly repetitive sequences based on resolution-enhanced 5D experiments. J Biomol NMR 48(3):169–177
Mukrasch M, Bibow S, Korukottu J, Jeganathan S, Biernat J, Griesinger C, Mandelkow E, Zweckstetter M (2009) Structural polymorphism of 441-residue Tau at single residue resolution. PLoS Biol 7(2):399–414
Narayanan RL, Durr UHN, Bibow S, Biernat J, Mandelkow E, Zweckstetter M (2010) Automatic assignment of the intrinsically disordered protein Tau with 441-residues. J Am Chem Soc 132(34):11906–11907
Nietlispach D, Ito Y, Laue ED (2002) A novel approach for the sequential backbone assignment of larger proteins: selective intra-HNCA and DQ-HNCA. J Am Chem Soc 124:11199–11207
Nováček J, Zawadzka-Kazimierczuk A, Papoušková V, Žídek L, Koźmiński W, Sklenář V (2011) 5D 13C-detected experiments for backbone assignment of unstructured proteins with a very low signal dispersion. J Biomol NMR 50(1):1–11
Orekhov VY, Ibraghimov IV, Billeter M (2001) MUNIN: a new approach to multi-dimensional NMR spectra interpretation. J Biomol NMR 20(1):49–60
Orekhov VY, Jaravine VA (2011) Analysis of non-uniformly sampled spectra with multi-dimensional decomposition. Prog Nucl Magn Reson 59(3):271–292
Panchal SC, Bhavesh NS, Hosur RV (2001) Improved 3D triple resonance experiments, HNN and HN(C)N, for H-N and N-15 sequential correlations in (C-13, N-15) labeled proteins: application to unfolded proteins. J Biomol NMR 20(2):135–147
Pannetier N, Houben K, Blanchard L, Marion D (2007) Optimized 3D-NMR sampling for resonance assignment of partially unfolded proteins. J Magn Reson 186(1):142–149
Permi P (2002) Intraresidual HNCA: an experiment for correlating only intraresidual backbone resonances. J Biomol NMR 23(3):201–209
Pervushin K, Vogeli B, Eletsky A (2002) Longitudinal H-1 relaxation optimization in TROSY NMR spectroscopy. J Am Chem Soc 124(43):12898–12902
Peterson FC, Deng Q, Zettl M, Prehoda KE, Lim WA, Way M, Volkman BF (2007) Multiple WASP-interacting protein recognition motifs are required for a functional interaction with N-WASp. J Biol Chem 282(11):8446–8453
Peti W, Smith LJ, Redfield C, Schwalbe H (2001) Chemical shifts in denatured proteins: resonance assignments for denatured ubiquitin and comparisons with other denatured proteins. J Biomol NMR 19(2):153–165
Rovnyak D, Frueh DP, Sastry M, Sun ZYJ, Stern AS, Hoch JC, Wagner G (2004) Accelerated acquisition of high resolution triple-resonance spectra using non-uniform sampling and maximum entropy reconstruction. J Magn Reson 170(1):15–21
Salmon L, Nodet G, Ozenne V, Yin G, Jensen MR, Zweckstetter M, Blackledge M (2010) NMR characterization of long-range order in intrinsically disordered proteins. J Am Chem Soc 132:8407–8418
Sasahara Y, Rachid R, Byrne MJ, de la Fuente MA, Abraham RT, Ramesh N, Geha RS (2002) Mechanism of recruitment of WASP to the immunological synapse and of its activation following TCR ligation. Mol Cell 10(6):1269–1281
Sattler M, Schleucher J, Griesinger C (1999) Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Mang Reson Spect 34(2):93–158
Schmieder P, Stern AS, Wagner G, Hoch JC (1994) Improved resolution in triple-resonance spectra by nonlinear sampling in the constant-time domain. J Biomol NMR 4(4):483–490
Sørensen OW, Eich GW, Levitt MH, Bodenhausen G, Ernst RR (1984) Product operator formalism for the description of NMR pulse experiments. Prog Nucl Mang Reson Spect 16:163–192
Stanek J, Koźmiński W (2002) Iterative algorithm of discrete Fourier transform for processing randomly sampled NMR data sets. J Biomol NMR 47(1):65–77
Stern AS, Li KB, Hoch JC (2002) Modern spectrum analysis in multidimensional NMR spectroscopy: comparison of linear-prediction extrapolation and maximum-entropy reconstruction. J Am Chem Soc 124(9):1982–1993
Sun ZYJ, Frueh DP, Selenko P, Hoch JC, Wagner G (2005) Fast assignment of N-15-HSQC peaks using high-resolution 3D HNcocaNH experiments with non-uniform sampling. J Biomol NMR 33(1):43–50
Szyperski T, Wider G, Bushweller JH, Wütrich K (1993) Reduced dimensionality in triple-resonance NMR experiments. J Am Chem Soc 115(20):9307–9308
Volkman BF, Prehoda KE, Scott JA, Peterson FC, Lim WA (2002) Structure of the N-WASP EVH1 domain-WIP complex: insight into the molecular basis of Wiskott-Aldrich Syndrome. Cell 111(4):565–576
Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT (2004) Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol 337(3):635–645
Wen J, Wu J, Zhou P (2011) Sparsely sampled high-resolution 4-D experiments for efficient backbone resonance assignment of disordered proteins. J Magn Reson 209(1):94–100
Yao J, Chung J, Eliezer D, Wright PE, Dyson HJ (2001) NMR structural and dynamic characterization of the acid-unfolded state of apomyoglobin provides insights into the early events in protein folding. Biochemistry 40(12):3561–3571
Zawadzka-Kazimierczuk A, Kazimierczuk K, Koźmiński W (2010) A set of 4D NMR experiments of enhanced resolution for easy resonance assignment in proteins. J Magn Reson 202(1):109–116
Zweckstetter M, Bax A (2001) Single-step determination of protein substructures using dipolar couplings: aid to structural genomics. J Am Chem Soc 123(39):9490–9491
Acknowledgments
This work was supported by the project "CEITEC - Central European Institute of Technology" from European Regional Development Fund, grant number CZ.1.05/1.1.00/02.0068, (J. N., L. Z., and V. S.) and by the Czech Science Foundation, grant numbers P206/11/0758 (J. N., L. Z., and V. S.). J.H.C. acknowledges the support of a Legacy Heritage personal grant by the Israel Science Foundation. Financial support by the Access to Research Infrastructures activity in the 7th Framework Programme of the EC (Contract 228461, EAST-NMR) for conducting the research is gratefully acknowledged. The project is a part of Joint Research Activity in the 7th Framework program of the EC (BioNMR n. 261863).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nováček, J., Haba, N.Y., Chill, J.H. et al. 4D Non-uniformly sampled HCBCACON and 1 J(NCα)-selective HCBCANCO experiments for the sequential assignment and chemical shift analysis of intrinsically disordered proteins. J Biomol NMR 53, 139–148 (2012). https://doi.org/10.1007/s10858-012-9631-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-012-9631-8