Abstract
A Citrus clementina gene, CcGASA4, which is involved in the responses of citrus to stress, was characterized. The gene was induced by Citrus tristeza virus infection, wounding and gibberellic, salicylic and abscisic acid treatments. A qRT-PCR analysis showed that CcGASA4 had a very high basal expression in flowers yet was still able to be further induced independently in giberellic, salicylic and abscisic acid-treated flowers. Subcellular localization analysis revealed that the CcGASA4 protein localized to the cell membrane and nucleus. A yeast two-hybrid analysis and bimolecular fluorescence complementation (BiFC) assays showed that CcGASA4 interacted with two proteins, the large proline-rich protein bag6-A (PRPBAG6-A) and the general negative regulator of transcription subunit 3 (CNOT3). PRPBAG6 has been reported to be involved in disease resistance. Replacing some of CcGASA4's conserved cysteines with alanines (Cys → Ala) abolished the protein’s interaction with CNOT3 but did not show any effect on the protein’s interaction with PRPBAG6-A. Thus, CcGASA4 appears to play multiple roles in Citrus, probably by interacting with different proteins and/or by localizing to different subcellular compartments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cysteine-rich peptides are particularly well represented among plants and are categorized into different classes based on the number and arrangement of the cysteine residues in the primary sequence (Silverstein et al. 2007). One class of cysteine-rich peptides, the gibberellic acid (GA) stimulated Arabidopsis (GASA)/GA-stimulated transcript (GAST) family plays crucial roles in plant growth, development and responses to abiotic and biotic stresses (Nahirnak et al. 2012a, b; Silverstein et al. 2007). Most GASA genes are primarily regulated by GA. GAST1 was the first GASA family gene identified in planta (Shi et al. 1992). GAST1 homologs have since been identified in different plant species, including GIP1-5 in petunia (Ben-Nissan et al. 2004), SN1–2 in potato (Segura et al. 1999; Berrocal-Lobo et al. 2002), FBCBP in French bean (Bindschedler et al. 2006), GASA1–15 in Arabidopsis thaliana (Herzog et al. 1995; Nahirnak et al. 2012a, b), FsGASA4 in beechnut (Alonso-Ramírez et al. 2009), ZmGSL1–10 in maize (Zimmermann et al. 2010), GsGASA1 in soybean (Li et al. 2011), CaSn in pepper (Mao et al. 2011). GASA proteins possess a cleavable signal peptide, consisting of 18–23 amino acid residues at the N-terminus, and a conserved GASA domain, consisting of 12 cysteines at the C terminus. However, a few GASA proteins such as PRGL in Gerbera hybrid (Peng et al. 2008) and AtGASA14 in Arabidopsis (Sun et al. 2013) contain an additional proline-rich protein (PRP) domain. GASA proteins also feature cysteine-disulfide bridges in their distinct three-dimensional structural folds (Silverstein et al. 2007). These disulfide bonds may be essential for the interactions of GASA proteins with other proteins (Nahirnak et al. 2012a, b).
The GASA4 gene was first reported in Arabidopsis (Herzog et al. 1995). It is GA-responsive and involved in plant reproduction. The expression pattern of AtGASA4 and the phenotypes of both the loss-of-function and the gain-of-function mutants clearly showed its involvement in bolting, branching, flowering and seed development in Arabidopsis (Rubinovich and Weiss 2010). The expression levels of GA metabolic genes and flowering-related genes were elevated in the Arabidopsis gasa4 mutant (Chen et al. 2007). Following exogenous GA treatments, AtGASA4 was up-expressed in meristematic tissues and young flower buds and was down-regulated in cotyledons, leaves, elongated roots, stems, petioles and floral organs (Aubert et al. 1998). An analysis of GASA4 mutants indicated that GASA4 regulates floral meristem identity and positively affects both seed size and total seed yield (Roxrud et al. 2007).
In addition, GASA4 shows a broad-spectrum anti-stress activity. Overexpression of wild-type GASA4 reduced the accumulation of reactive oxygen species, while the cysteine to alanine mutant inhibited H2O2 reduction (Rubinovich and Weiss 2010). Overexpressing the maize GASA4 enhanced the heat tolerance of Arabidopsis (Ko et al. 2007). Overexpressing the Fagus sylvatica FsGASA4 increased tolerance to salt, oxidative and heat stresses, and increased the endogenous salicylic acid (SA) content in seeds of Arabidopsis (Alonso-Ramírez et al. 2009).
The GASA proteins’ N-terminal signal peptide suggests that these proteins may be secretory. N-terminal signal peptides must be cleaved to allow the proteins to localize accurately in cells (Qu et al. 2016). Different GASA proteins may differ in subcellular localizations. For example, GIP2 and GIP5 are localized in the cell wall, while GIP1 and GIP4 are confined to the endoplasmic reticulum membrane in petunia (Ben-Nissan et al. 2004). Soybean (Glycine soja) GsGASA1 can localize to different subcellular compartments, including cell wall, cytoplasm and nucleus (Li et al. 2011).
However, little is known about the role of GASA4 in plant responses to biotic stresses caused by pathogens. In our previous gene chip experiments, we found that the citrus homolog of AtGASA4, Cit.11064.1.S1_at, was highly induced in citrus leaves after infection with Citrus tristeza virus (CTV) (Zhang 2010), indicating that GASA4 may also play a role in biotic stress responses. In this report, we show that CcGASA4 is indeed responsive to CTV infection.
Materials and methods
Plant materials and growth conditions
Citrus plants (Citrus clementina and Citrus sinensis cv. Jincheng) were grown in the greenhouse of the Institute of Fruit Tree Research, Guangdong Academy of Agricultural Sciences. Arabidopsis thaliana plants, ecotype Columbia Col-0, were used for transient expression and a protein–protein interaction analysis, and were grown at 22 °C under 16 h light/8 h dark photocycle. Anion epidermal cells were used for analyzing protein localizations.
Hormone treatments and the growth assay
For the CTV treatment, ten Citrus sinensis seedlings were randomly assigned into two groups, each containing five seedlings. The treatment and the control groups were graft-inoculated with CTV virulent strain CT14-infected and with virus-free Citrus sinensis branches, respectively. Leaf samples were taken at 1, 3 and 5 months after treatment. For hormone treatments, 200 mg/L GA3, 500 μΜ SA, 100 μM indole-3-acetic acid (IAA) and 200 μΜ abscisic acid (ABA) (Reckova et al. 2019) were independently sprayed onto Jincheng leaves and flowers, with water as the control. For mechanical injury, Jincheng leaves were pressed with forceps until clear dents were seen on the surfaces. Leaves were collected at 1, 4, 12 and 24 h, respectively, while the flowers were collected at 2, 4, 8 and 16 h after treatment. For the tissue-specific expression analysis, samples of roots, stems, leaves, fully developed flowers and newly opened flowers were collected.
RNA and DNA extraction
Total RNA was extracted using an RNA extraction kit (Bioteke, China) following the manufacturer’s instructions. Genomic DNA was extracted using a Genomic DNA Rapid Extraction Kit (Dongsheng Biotech, China) following the user’s manual. The quality and quantity of total RNA and DNA were analyzed on a NanoDrop 2000C (Thermo Fisher Scientific, USA).
CcGASA4 cloning
All primers used in this study were synthesized by Beijing Liuhe Huada Gene Technology Company (China) (see Table S1 in Supplementary Material). cDNA was synthesized from total leaf RNA of Citrus clementina using a PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa, Japan). The complete coding sequence of CcGASA4 (GenBank accession number: MN781588) was amplified by PCR from cDNA using the primer pair GASA4-F/R. The PCR was performed on an ABI thermal cycler (Thermo Fisher Scientific, USA). PCR products were purified using an AxyPrep™ DNA Gel Extraction Kit (Axygen, USA), and they were then independently cloned into the pMD®20-T vector (TaKaRa). The same primers and PCR protocol were also used to amplify the gene from genomic DNA. PCR products were directly sequenced. All the sequencing was conducted by the Sangon Biotechnology Company (China).
Bioinformatics analysis
The CcGASA4 gene sequence (Ciclev10013454m CDS) was downloaded from the Phytozome (https://www.phytozome.org). cDNA and genomic DNA sequence analysis and the sequence alignment of the homologous proteins were conducted in DNAMAN5.2.2. The protein secondary structure prediction was performed using YASPIN (https://www.ibi.vu.nl/programs/yaspinwww/). The signal peptide and GASA domain were identified using InterProScan (https://www.cbs.dtu.dk/services/SignalP/; https://www.ebi.ac.uk/Tools/pfa/iprscan/). The phylogenetic tree was generated for GASA4 and its homologs using the Neighbor-joining method in MEGA 5.0. The cis-acting elements were identified from the CcGASA4 promoter using PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al. 2002).
Gene expression analysis
The CcGASA4 expression was quantified using qRT-PCR with gene-specific primers and the reference gene β-actin primers. qRT-PCR reactions were performed on a Bio-RAD CFX Connect™ Real-Time System (Bio-Rad, USA) using SYBR®Premix Ex Taq™ II (Perfect Real Time) (TaKaRa, Japan). The expression ratios of the analyzed gene over the reference gene were determined by calculating the 2−ΔΔCt.
Subcellular localization
cDNA was amplified using primers GASA4–GFPF/R, digested with XbaI and BamHI (TaKaRa, Japan), and cloned into the pBEGFP vector to generate the CcGASA4–GFP fusion construct. Onion epidermal cells were transformed independently with the construct and the control original pBEGFP vector by agroinfiltration (Sun et al. 2007) and incubated in the dark for 36 h. The GFP fluorescence was then observed under a confocal laser scanning microscope.
Yeast two-hybrid (Y2H) assay
A yeast cDNA library was prepared using Clontech’s Make Your Own “Make & Plate™” Library System (Clontech, USA). Total RNA was extracted from leaves of Citrus sinensis trees infected with Candidatas Liberibacter asiaticus (CLas), a bacterium causing the disease Huanglongbing, and used to generate cDNA. cDNA amplification was performed using long distance PCR (SMART™ technology, Clontech). The purified cDNA and the pGADT7-Rec linearized vector were co-transformed into Y187 yeast cells.
The open reading frame (ORF) of CcGASA4 was amplified using the primers GASA4BAIT-F/R, digested with EcoRI and BamHI (TaKaRa, Japan), and cloned into pGBKT7 (Clontech, USA) to produce the bait vector pGBKT7–GASA4. Preparation of Y2HGold yeast competent cells and the subsequent transformation of the cells with pGBKT7–GASA4 plasmid were performed by following the same protocol used in the yeast cDNA library preparation. The pGBKT7–GASA4 vector without autoactivation activity and toxicity in the yeast cells was selected as a bait to screen the citrus cDNA library for proteins interacting with GASA4 by following the instructions of the Matchmaker™ Gold Yeast Two-Hybrid System (Clontech, USA). The candidate CcGASA4-interacting protein genes were cloned and inserted into pGADT7 vectors to verify their interactions with GASA4 using the yeast two-hybrid assay.
Bimolecular fluorescent complementary (BiFC) analysis
The GASA4 cDNA was inserted into pSAT4–nEYFP, and the cDNAs of the CcGASA4 interacting proteins were individually inserted into the pSAT4–cEYFP vector using the ClonExpress II One Step Cloning Kit (Vazyme, China). All the constructed BiFC plasmids were then propagated in E. coli, extracted using Magen’s HiPure Plasmid EF Mini Kit, and transformed into Arabidopsis protoplast cells. The GASA4–GFP plasmid was used as a control to verify if the transformation was successful. The transformed protoplasts were incubated at room temperature for 16 h in darkness as described previously (Wu et al. 2009). The florescence signals from transiently expressed GASA4–nEYFP and interacting-protein-cEYFPs were observed under a Zeiss LSM710 laser scanning confocal microscope.
Creation of cysteine-to-alanine mutations
The gene’s cysteine to alanine mutations were created by PCR by using primers that contained one or two alanine (GCG) codons at the sites corresponding to the original cysteine residues. Specifically, mutant 1 was mutated at residues 48, 52 and 56; mutant 2 was mutated at 72, 73, 76 and 78; and mutant 3 was mutated at 90, 92 and 105. The first round of PCR was performed using the CcGASA4 plasmid as a template to amplify the fragments 1, 2, 3, 4 and 5 (Fig. S1). The second round of overlapping PCR was as follows: mutant 1 was generated using fragments 1 and 2 as templates with GASA4BDF/R as primers; mutant 2 was created using fragments 3 and 4 as templates with GASA4–BDF/R as primers; and mutant 3 was created using fragment 5 as the template with GASA4–BDF and GASA4–T3R2 as primers.
Results
Features of the CcGASA4 gene and it’s deduced protein
Both the cDNA and the genomic DNA sequences of CcGASA4 were cloned from clementine mandarin leaves using the gene specific primers GASA4F and GASA4A (Fig. 1a). The cDNA contained a 321 bp-long ORF which shared a similarity of 99.69% to the clementine mandarin gene Ciclev10013454m CDS (https://www.phytozome.org). The cloned genomic DNA contained four exons and three introns (Fig. S2), and it was essentially identical to that of Ciclev10013454m.g.
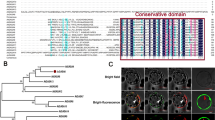
Cloning of the CcGASA4 gene and bioinformatics analysis of its deduced amino acid sequence. a PCR amplifications of CcGASA4 from genomic DNA and cDNA. M is DS™ 2000 marker. b Phylogenetic tree showing the similarities between the deduced amino acid sequence of CcGASA4 and its homologs from other species. The black triangle represents the CcGASA4 sequence. The homologs are as follows: Vitis vinifera (XP_002283517.1), Cicer arietinum (XP_004491797.1), Gossypium hirsutum (AGE15501.1), Fagus sylvatica (CAJ77893.1), Citrus clementina (XP_006431148.1), Cucumis sativus (XP_004147472.1), Gerbera hybrid cultivar (AGL92232.1), Solanum lycopersicum (XP_004236040.1), Petunia hybrid (CAD10106.1), Rosa damascene (ACM47319.1), Fragaria vesca subsp. Vesca (XP_004304306.1) and Arabidopsis thaliana (AAG52379.1). c Multiple alignment of CcGASA4 and its homologs from other species. Lowercase letters indicate the conserved residues. Different background colors highlight residues having different conservation levels. The underline indicates the 60-amino acid GASA domain. d Partial sequence alignment of GASA4 and the three mutants created in the study. The numbers indicate the sites where the original cysteines were replaced by alanines
The deduced peptide sequence was 106 aa and contained a signal peptide, a transmembrane domain and a GASA domain (Fig. 1b). A BLAST alignment showed that the similarities between the predicted CcGASA4 protein and the GAST-like proteins from cucumber, grape, cotton and Rosa damascene were greater than 60%. A multiple sequence alignment showed that the C-terminal was much more conserved than the N-terminal (Fig. 1b), with no changes existing in the 12 cysteine residues of the C-terminal among all the analyzed GASA4 homologous proteins. Two subclades were clearly identifiable on the phylogenetic tree (Fig. 1c): the first included Arabidopsis, Rosa damascene and strawberry, and the second included the CcGASA4 protein cloned in this study and the remaining analyzed GAST-like proteins. CcGASA4 was evolutionally closer to a cucumber homolog.
CcGASA4 responds to stresses and hormones in Citrus leaves
The effects of CTV infection on the expression of GASA4 were investigated in leaves of Jincheng. GASA4 was up-regulated at 1 month post-CTV inoculation. The peak expression was observed after 3 months. Although it decreased substantially, the gene’s expression was still higher in CTV-infected leaves than in control leaves after 5 months (Fig. 2a). The CcGASA4 promoter sequence downloaded from Phytozome was 1800 bp. It contains the gibberellin response element GARE, the abscisic acid response element ABRE, the inductive salicylic acid and the damage signal element WBox, the inductive pathogen invasion and the salt stress element GT1- Box, the drought response element MYB, the MYC and MYB transcription factor binding site MBS, the pollen-specific activation element AGAAA, the auxin response factor binding site AFR, and the photo-responsive elements, GBox, Box4 and IBox core (Fig. S3). Thus, CcGASA4 might participate in hormone and stress responses. Therefore, hormone and wounding treatments were performed on Jincheng leaves. As shown in Fig. 2b, the gene was significantly induced after 4 h in leaves treated with GA3, and a small peak induction was shown at 12 h. Following the SA treatment, CcGASA4 expression increased at 1 h and reached a peak of 87-fold increase at 4 h, as compared with the control. In ABA treatment, CcGASA4 expression showed two peaks, the first one at 4 h and the second one at 24 h, corresponding to increases of 12.9-fold and 17.8-fold, respectively. CcGASA4 was also up-regulated by mechanical injury with a peak expression of 6.21-fold after 24 h.
Expression levels of CcGASA4 in leaves and flowers. a The expression of GASA4 in leaves of CTV-infected Jincheng seedlings after 1, 3 and 5 months of treatment. b qRT-PCR analysis of CcGASA4 expression in leaves of Jincheng seedlings treated with GA, SA and ABA as well as after wounding. c Expression of CcGASA4 in roots, stems, leaves and flowers. d Expression level of CcGASA4 in flowers at different flowering stages: sprout stage, S1; flower-bud stage, S2; petal-display stage, S3; initiating bloom stage, S4; bloom stage, S5; and wither stage, S6. eCcGASA4 expression in different floral tissues (anthers, petals, filaments and styles). f Effects of GA, SA, ABA and IAA treatments on CcGASA4 expression in flowers. All the experiments were replicated three times. Bars indicate SEs
CcGASA4 is highly expressed in flowers
The variations in CcGASA4 expression were investigated in Citrus roots, stems, leaves and flowers. Figure 2c showed that the gene was expressed, although at different levels, in all the analyzed tissues. Notably, the gene was highly abundantly expressed in flowers. The gene’s temporal expression pattern was investigated in flowers, and it was expressed higher at the initiating bloom and bloom stages, with the lowest expression at the wither stage (Fig. 2d). As shown in Fig. 2e, the gene varied in its expression level in different flower parts, with the highest expression in petals and the lowest in anthers.
GA, SA and ABA promote GASA4 expression in flowers
With the GA treatment, the expression of GASA4 spiked after 2 h and then subsided (Fig. 2f). The SA treatment gradually stimulated the gene’s expression in the first 4 h and then reduced its expression in the following hours. ABA increased the expression of GASA4 to a peak at 2 h but not after 8 h. Interestingly, although its promoter contains an auxin response factor binding AFR site, the gene’s expression was not induced by the IAA treatment.
CcGASA4 localizes to the plasma membrane and nucleus
The CcGASA4-GFP was successfully constructed (Fig. 3a) and transiently expressed in onion epidermal cells (Fig. 3b). After plasmolysis, GFP fluorescence signals were observed in the nucleus and plasma membrane but not in the cell wall.
Subcellular localization of CcGASA4-GFP protein in onion epidermal cells. a The schematic illustration of the two vectors: PBEGFP and CcGASA4-GFP. The cauliflower mosaic virus 35S promoter was used to drive the transient expression of the CcGASA4-GFP fusion protein in Arabidopsis protoplasts. b Fluorescence images showing the subcellular locations of the fusion proteins. (1) Hoechst 33,342 staining; (2) fluorescent field; (3) bright field; and (4) merged. Scale bar = 100 μm.
CcGASA4 interacts with two different proteins in Y2H system
A representative pGADT7-DEST-cDNA yeast library was constructed using mRNA from leaves of sweet orange trees infected with CLas bacteria. The pGBKT7-GASA4 bait vector was constructed and successfully transformed into Y2HGold yeast. In total, 12 positive clones were obtained after screening for GASA4-interacting proteins on the auxotrophic medium DDO/X/A and the chromogenic medium QDO/X/A. Plasmids from all the positive clones were extracted (Table S2 in Supplementary Material), transformed into E. coli and sequenced. The yeast two-hybrid assay showed that only two of the proteins PRPBAG6-A and CNOT3 interacted with CcGASA4 (Fig. 4a). In the BiFC assay, green florescence signals were detected on cell membranes and nuclei following co-transformation of CcGASA4 with either CNOT3 or PRPBAG6-A (Fig. 4b) further verifying the interactions of CcGASA4 with PRPBAG6-A and with CNOT3.
Interactions of CcGASA4 with both PR6PBAG6-A and CNOT3. a Identification of GASA4 interacting proteins by Y2H assay. The GAL4 DNA-binding domain was fused to the CcGASA4 coding region, forming BD-GASA4, and the three mutated CcGASA4 sequences, BD-mut1, BD-mut2 and BD-mut3, were used as bait vectors. The cDNA sequences of PRPBAG6-A and CNOT3 isolated from the citrus cDNA library were fused with the Gal4 activation domain to construct the prey vectors, AD-BAG6 and AD-CNOT3, respectively. Yeast was grown on QDO/X/A (SD/–Ade /–His/–L eu/–rp/x-α-gal/Aba) medium. b BiFC verification of protein interactions. The transient expression of the BiFC plasmids in Arabidopsis leaf protoplasts are shown by individual and merged images of GFP and chlorophyll autofluorescence (Chl), as well as DIC images, of protoplasts. Scale bars = 5 μm. a GFP vector; b GASA4-GFP vector; c large proline-rich protein bag6-A (PRPBAG6-A)-GFP vector; d general negative regulator of transcription subunit 3 (CNOT3)-GFP vector; e CcGASA4-nYFP vector + PRPBAG6-A-cYFP vector; and f CcGASA4-nYFP vector + CNOT3-cYFP vector.
Mutations in specific cysteine residues abolish CcGASA4′s interaction with CNOT3
To investigate if the cysteine residues participate in the interactions of CcGASA4 with PRPBAG6-A and CNOT3, three CcGASA4 constructs, each containing 3–4 cysteine to alanine (C → A) mutations (Fig. 1d) were made and subjected to yeast two-hybrid assay. As shown in Fig. 4a, these mutants were still capable of interacting with PRPBAG6-A but not with CNOT3.
Discussion
We previously found that a GASA4-like protein gene (Cit.11064.1.S1_at) was highly up-regulated by CTV-infection in Jincheng (Zhang 2010). In this study, we isolated, sequenced and characterized the gene designated as CcGASA4. We showed that the gene was responsive to biotic and abiotic stresses and stress-related hormones in citrus, and demonstrated that the protein interacted with PRPBAG6-A and CNOT3 and that its interaction with CNOT3 was associated with cysteine residues. Results showed that CcGASA4 was localized to the plasma membrane and the nuclei.
GASA genes are responsive to GAs (Sun et al. 2013). Although the basal GASA4/GUS activity was only slightly enhanced in root and flower meristems of the wild type Arabidopsis by the GA treatment, the GA-dependent expression of GASA4 was evident in GA-deficient mutants (Aubert et al. 1998). Our study demonstrated that CcGASA4 was up-regulated in flowers and leaves of citrus by an exogenous GA treatment, indicating that CcGASA4 is also GA-responsive. The involvement of GA in flowering has been well documented in higher plants. That the CcGASA4 expression was much higher in flowers than in other organs may signify its importance in flower development. GASA6, a GASA4 homolog in Arabidopsis was reported to affect flowering time as indicated by the GASA6 over-expression plants being early-flowering and the gasa4/gasa6 double mutants being late-flowering (Qu et al. 2016).
The presence of multiple stress-responsive elements in CcGASA4′s promoter suggests it may be a stress-responsive gene. Treatments with the stress-related hormones SA and ABA, as well as wounding, up-regulated CcGASA4 in Citrus. Similarly, the rice GAST family gene OsGASR1 was induced by salt and ABA treatments, and over-expressing OsGASR1 enhanced salt tolerance (Lee et al. 2015). Its stress responsiveness may involve cysteine residues because over-expressing the GAST-like gene GIP2 reduces H2O2 levels in wound-treated petunia leaves and in osmotic stress- and ABA-treated guard cells (Wigoda et al. 2006). Rubinovich and Weiss (2010) showed that over-expressing GASA4 suppressed ROS accumulation in Arabidopsis and that the transgenic seeds were partially resistant to NO donor sodium nitroprusside. Additionally, E. coli cells expressing either an intact GASA4 or a truncated version containing only the cysteine-rich domain were resistant to sodium nitroprusside. They also showed that the gasa4 with four C → A mutations lost redox activity and GA responsiveness, suggesting that the two functions are interlinked.
We showed that the citrus GASA4 gene was induced by CTV infection. This is the first report, to our knowledge, that GASA4 is also responsive to viral infection. The potato Snakin-1 has in vivo antibacterial activity (Segura et al. 1999), and Snakin-2 has both anti-bacterial and anti-fungal activities (Berrocal-Lobo et al. 2002). The pepper CaSn protein has an anti-nematode activity (Mao et al. 2011). A compound protein containing a French bean Snakin-2-like protein has antimicrobial activity (Bindschedler et al. 2006).
Two CcGASA4-binding proteins, PRPBAG6-A and CNOT3, were identified in this study (Fig. 4b). The French bean Snakin-2-like protein can form a two-component protein complex with a PRP to bind the pathogen Colletotrichum lindemuthianum (Bindschedler et al. 2006). We, therefore, hypothesize that the interaction between CcGASA4 and PRPBAG6-A, also a PRP, may be similarly involved in defense in Citrus. CNOT3 is a component of the CCR4–NOT complex, an essential and conserved multi-subunit complex that plays multiple roles, including suppressing tumors in drosophila and in humans, maintaining self-renewal program in ES cells, and regulating RNA turnover and transcription termination (Vicente et al. 2018; Martufi 2014). Because CNOT3 interacts with Nucleolin and Histone H1 (Martufi 2014), GASA4′s nuclear localization and its interaction with CNOT3 may signify its potential role in chromatin organization.
Cysteine is required for the formation of disulfide bonds inside a single peptide or between different subunits of a compound protein, and thus, it plays an important role in stabilizing the protein’s spatial conformation. The 12 conserved cysteine residues present in all the GASA proteins, including the citrus CcGASA4, can theoretically form 5 to 6 disulfide bonds (Ben-Nissan et al. 2004). Although the replacement of cysteine residues with alanines impaired the protein’s GA responses and redox activity (Rubinovich and Weiss 2010), no reports have demonstrated roles for cysteine residues in the interactions between GASA4 and other proteins. In this study, we showed that the interaction between the C → A mutated gasa4 protein and CNOT3 was totally abolished but not the interaction between gasa4 and PRPBAG6-A. This may indicate that the interaction between GASA4 and CNOT3 requires correctly-folded GASA4, which requires disulfide bonds, whereas the interaction between GASA4 and PRPBAG6-A may involve only limited surface areas of GASA4. Clearly, the binding sites of GASA4 to PRPBAG6-A and CNOT3 are different.
Conclusions
Previous reports and this study showed that GASA4 is involved not only in plant growth and development but also in biotic and abiotic stress responses. Clues to why it plays diverse roles are partially provided by this study, because it resides in different subcellular compartments, and/or it interacts with different proteins. However, the exact mechanisms are still elusive and further studies are required.
References
Alonso-Ramírez A, Rodríguez D, Reyes D, Jiménez JA, Nicolás G, López-Climent M, Gómez-Cadenas A, Nicolás C (2009) Evidence for a role of gibberellins in salicylic acid-modulated early plant responses to abiotic stress in arabidopsis seeds. Plant Physiol 150:1335–1344
Aubert D, Chevillard M, Dorne AM, Arlaud G, Herzog M (1998) Expression patterns of GASA genes in Arabidopsis thaliana: the GASA4 gene is up-regulated by gibberellins in meristematic regions. Plant Mol Biol 36:871–883
Ben-Nissan G, Lee J, Borohov A, Weiss D (2004) GIP, a Petunia hybrida GA-induced cysteine-rich protein: a possible role in shoot elongation and transition to flowering. Plant J 37:229–238
Berrocal-Lobo M, Segura A, Moreno M, Lopez G, Garciaolmedo F, Molina A (2002) Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiol 128:951–961
Bindschedler LV, Whitelegge JP, Millar DJ, Bolwell GP (2006) A two component chitin-binding protein from French bean—association of a proline-rich protein with a cysteine-rich polypeptide. FEBS Lett 580:1541–1546
Chen I, Lee S, Pan S, Hsieh H (2007) GASA4, a GA-stimulated gene, participates in light signaling in Arabidopsis. Plant Sci 172:1062–1071
Herzog M, Dorne AM, Grellet F (1995) GASA, a gibberellin-regulated gene family from Arabidopsis thaliana related to the tomato GAST1 gene. Plant Mol Biol 27:743–752
Ko C, Woo Y, Lee DJ, Lee M, Kim CS (2007) Enhanced tolerance to heat stress in transgenic plants expressing the GASA4 gene. Plant Physiol Biochem 45:722–728
Lee S, Han S, Kim S (2015) Salt- and ABA-inducible OsGASR1 is involved in salt tolerance. J Plant Biol 58:96–101
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, De Peer YV, Rouz P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Li KL, Bai X, Li Y, Cai H, Ji W, Tang LL, Wen YD, Zhu YM (2011) GsGASA1 mediated root growth inhibition in response to chronic cold stress is marked by the accumulation of DELLAs. J Plant Physiol 168:2153–2160
Mao Z, Zheng J, Wang Y, Chen G, Yang Y, Feng D, Xie B (2011) The new CaSn gene belonging to the snakin family induces resistance against root-knot nematode infection in pepper. Phytoparasitica 39:151–164
Martufi M (2014) Role of Cnot3 in gene regulation and cell cycle progression. Doctoral dissertation, Imperial College, London
Nahirnak V, Almasia NI, Fernandez PV, Hopp HE, Estevez JM, Carrari F, Vazquez-Rovere C (2012a) Potato snakin-1 gene silencing affects cell division, primary metabolism, and cell wall composition. Plant Physiol 158:252–263
Nahirnak V, Almasia NI, Hopp HE, Vazquezrovere C (2012b) Snakin/GASA proteins involvement in hormone crosstalk and redox homeostasis. Plant Signal Behav 7:1004–1008
Peng JZ, Lai LJ, Wang XJ (2008) PRGL: A cell wall proline-rich protein containning GASA domain in Gerbera hybrida. Sci China Life Sci 51:520–525
Qu J, Kang SG, Hah C, Jang J (2016) Molecular and cellular characterization of GA-stimulated transcripts GASA4 and GASA6 in Arabidopsis thaliana. Plant Sci 246:1–10
Reckova S, Tuma J, Dobrev P, Vankova R (2019) Influence of copper on hormone content and selected morphological, physiological and biochemical parameters of hydroponically grown Zea mays plants. Plant Growth Regul 89(2):191–201
Roxrud I, Lid SE, Fletcher JC, Schmidt ED, Opsahlsorteberg H (2007) GASA4, one of the 14-member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol 48:471–483
Rubinovich L, Weiss D (2010) The Arabidopsis cysteine-rich protein GASA4 promotes GA responses and exhibits redox activity in bacteria and in planta. Plant J 64:1018–1027
Segura A, Moreno M, Madueño F, Molina A, García-Olmedo F (1999) Snakin-1, a peptide from potato that is active against plant pathogens. Mol Plant Microbe In 12:16–23
Shi L, Gast RT, Gopalraj M, Olszewski NE (1992) Characterization of a shoot-specific, GA3- and ABA-regulated gene from tomato. Plant J 2:153–159
Silverstein KA, Moskal WA, Wu HC, Underwood BA, Graham MA, Town CD, Vandenbosch KA (2007) Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J 51:262–280
Sun W, Cao Z, Li Y, Zhao Y, Zhang H (2007) A simple and effective method for protein subcellular localization using agrobacterium-mediated transformation of onion epidermal cells. Biologia 62:529–532
Sun S, Wang H, Yu H, Zhong C, Zhang X, Peng J, Wang X (2013) GASA14 regulates leaf expansion and abiotic stress resistance by modulating reactive oxygen species accumulation. J Exp Bot 64:1637–1647
Vicente C, Stirparo R, Demeyer S, de Bock CE, Gielen O, Atkins M, Yan JK, Halder G, Hassan BA, Cools J (2018) The CCR4-NOT complex is a tumor suppressor in Drosophila melanogaster eye cancer models. J Hematol Oncol 11:108
Wigoda N, Bennissan G, Granot D, Schwartz A, Weiss D (2006) The gibberellin-induced, cysteine-rich protein GIP2 from Petunia hybrida exhibits in planta antioxidant activity. Plant J 48:796–805
Wu F, Shen S, Lee L, Lee S, Chan M, Lin C (2009) Tape-Arabidopsis sandwich—a simpler Arabidopsis protoplast isolation method. Plant Methods 5:16–16
Zhang LY (2010) Transcriptome profiling of genes involved in ethylene-induced fruit abscission, cloning and characterization of ethylene-regulated genes in citrus sinensis. PhD thesis, Southwest University, China
Zimmermann R, Sakai H, Hochholdinger F (2010) The gibberellic acid stimulated-like gene family in Maize and Its role in lateral root development. Plant Physiol 152:356–365
Acknowledgements
This work was supported by the Key-Area Research and Development Program of Guangdong Province (2018B020202009), National Natural Sciences Foundation of China (Grant No. 3157111362), Guangdong Provincial Science and Technology Programs (2019B030316005, 2017A030310281), Guangdong Provincial Special Fund For Modern Agriculture Industry Technology Innovation Teams (2019KJ108), the Natural Science Foundation of China Chongqing (cstc2018jcyjAX0384) and Fujian Provincial Science and Technology Program (2017J01615). The authors would like to thank Dr. Minlun Hu for his experimental support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10725_2020_589_MOESM1_ESM.jpg
Supplementary Fig. S1—Creation of cysteine-to-alanine mutations. Segment 1 was amplified using primers of GASA4-BDF and GASA4-T1R; segment 2 was amplified using primers of GASA4-T1F and GASA4-BDR; segment 3 was amplified using primers of GASA4-BDF and GASA4-T2R; segment 4 was amplified using primers of GASA4-T2F and GASA4-BDR; and segment 5 was amplified using primers of GASA4-BDF and GASA4-T3R1. Mutant 1 was generated using fragments 1 and 2 as templates with GASA4BDF and GASA4BDR as primers; mutant 2 was created using fragments 3 and 4 as templates with GASA4-BDF and GASA4-BDR as primers; and mutant 3 was created using fragment 5 as the template with GASA4-BDF and GASA4-T3R2 as primers(JPG 1127 kb)
10725_2020_589_MOESM3_ESM.jpg
Supplementary Fig. S3—Analysis of cis-acting elements in the CcGASA4 promoter sequence. The underlined sequences represent the different cis-acting elements that are marked by their names. GARE, ABRE, WBox, GT1- Box, MBS and AFR are cis-acting elements responsive to gibberellin, abscisic acid, salicylic acid and wounding signals, pathogen invasion and salt, drought and auxin-related stresses, respectively. AGAAA is a pollen-specific activation element. (JPG 1672 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wu, T., Cheng, C., Zhong, Y. et al. Molecular characterization of the gibberellin-stimulated transcript of GASA4 in Citrus. Plant Growth Regul 91, 89–99 (2020). https://doi.org/10.1007/s10725-020-00589-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-020-00589-1