Abstract
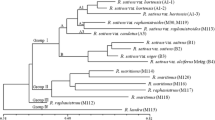
A local collection of 33Saccharum spontaneum L. clones and two sugarcane cultivars (LCP 82-89 and LCP 85-384) were assessed for genetic variability using random amplified polymorphic DNA (RAPD)-PCR. A total of 157 polymorphic RAPD-PCR bands were scored with 17 primers. The number of RAPD-PCR products per primer ranged from four to 16. The data were analyzed with two multivariate analysis software programs, NTSYSpc and DNAMAN®. Although these two programs yielded similar results, a bootstrapped phylogenetic tree could only be generated with the DNAMAN® software. A substantial degree of genetic diversity was found within the localS. spontaneum collection. Pairwise genetic homology coefficients ranged from 65% (SES, 196/Tainan 2n = 96) to 88.5% (IND 81-80/IND 81-144). LCP 82-89 and LCP 85-384 shared a greater similarity (82%) than either was to any clone ofS. spontaneum (ranging from 60.5 to 75.2%). The 33S. spontaneum clones were assigned to eight groups independent of their geographic origin or morphology, while the two sugarcane cultivars were assigned to the ninth group. All but two pairs ofS. spontaneum clones could be distinguished by a single RAPD primer OPBB-02. The use of a second primer, either OPBE-04 or Primer 262, separated allS. spontaneum clones. One amplification product from the RAPD primer OPA-11, OPA-11-336, proved to be cultivar-specific and has been adopted for use in our breeding program. Information from this study would help conserve the genetic diversity ofS. spontaneum.
Similar content being viewed by others
References
Arceneaux G. 1967. Cultivated sugarcanes of the world and their botanical derivation. Proc 12th Intl. Soc. Sugar Cane Technol., pp. 844–854.
Artschwager E. and Brandes E.W. 1958. Sugarcane (Saccharum officinarum L.): Origin, classification, characteristics and descriptions of representative clones. USDA Handbook No. 122.
Besse P., McIntyre C.L. and Berding B.N. 1997. Characterisation ofErianthus sect.Ripidium andSaccharum germplasm (Andropogoneae-Saccharinae) using RFLP markers. Euphytica 93: 283–292.
Besse P., Taylor G., Carroll B., Berding N., Burner D.M. and McIntyre C.L. 1998. Assessing genetic diversity in a sugarcane germplasm collection using an automated AFLP analysis. Genetica 104: 143–153.
Brandes E.W. 1958. Origin, classification and characteristics. In: Artschwager E. and Brandes E.W. (eds), Sugarcane (Saccharum officinarum L.), U.S. Dept. Agric. Handbook 122: 1–35, 260–262.
Burner D.M. and Legendre B.L. 1993. Sugarcane genome amplification for the subtropics: a 20-year effort. Sugar Cane 3: 5–10.
Burner D.M., Pan Y.-B. and Webster R.D. 1997. Genetic diversity of North American and Old WorldSaccharum assessed by RAPD analysis. Genet. Resour. Crop Evol. 44: 235–240.
Chu T.L., Juang P.Y. and Shang K.C. 1962. The wild cane (S. spontaneum) in Taiwan. Reporter Taiwan Expt. Station 28: 1–11.
Daniels J. and Roach B.T. 1987. Taxonomy and evolution. In: Heinz D.J. (ed.), Sugarcane Improvement Through Breeding. Elsevier, New York, pp. 7–84.
Dunckelman P.H. and Breaux R.D. 1969. Agronomic characteristics ofSaccharum spontaneum in culture in Houma, Louisiana. Intl. Sugar J. 71: 333–334.
Dunckelman P.H. and Legendre B.L. 1982. Guide To Sugarcane Breeding in the Temperate Zone. USDA-ARS, ARM-S-22, New Orleans.
Feng D.F. and Doolittle R.F. 1987. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J. Mol. Evol. 25: 351–360.
Fritsch P., Hanson M.A., Spore C.D., Pack P.E. and Rieseberg L.H. 1993. Constancy of RAPD primer amplification strength among distantly related taxa of flowering plants. Plant Mol. Biol. Rep. 11: 10–20.
Grassl C.O. 1946.Saccharum robustum and other wild relatives of ‘noble’ sugar canes. J. Arnold Arbor. 27: 234–252.
Grassl C.O. 1969.Saccharum names and their interpretation. Proc. Intl. Soc. Sugar Cane Technol. 13: 868–875.
Harvey M. and Botha F.C. 1996. Use of PCR-based methodologies for the determination of DNA diversity betweenSaccharum varieties. Euphytica 89: 257–265.
Harvey M., Huckett B.I. and Botha F.C. 1994. Use of polymerase chain reaction (PCR) and random amplification of polymorphic DNAs (RAPDs) for the determination of genetic distances between 21 sugarcane varieties. Proc. S. Afr. Sugar Technol. Assn. 68: 36–40.
Huckett B.I. and Botha F.C. 1995. Stability and potential use of RAPD markers in a sugarcane genealogy. Euphytica 86: 117–125.
Jannoo N., Grivet L., Seguin M., Paulet F., Domaingue R., Rao P.S., Dookun A., D’Hont A. and Glaszmann J.C. 1999. Molecular investigation of the genetic base of sugarcane cultivars. Theor. Appl. Genet. 99: 171–184.
Kandasami P.A., Sreenivasan T.V., Palanichami K. and Ramana Rao T.C. 1983. Sugarcane germplasm: classification of clones. Sugar Cane 2: 1–3.
Linnaeus C. 1753. Species plantarum, vol. 2, Stockholm. In: 1959 Facsimile Edition, Ray Society, London.
Linnaeus C. 1771. Mantissa plantarum altera. Stockholm. In: 1961 Facsimile Edition, J. Cramer, Weinheim, p. 183.
Legendre B.L. and Breaux R.D. 1983. The USDA basic sugarcane breeding program in Louisiana. Proc. Inter-Amer. Sugar Cane Sem.: Varieties and Breeding III: 96–98.
Lu Y.H., D’Hont A., Paulet F., Grivet L., Arnaud M. and Glaszmann J.C. 1994. Molecular diversity and genome structure in modern sugarcane varieties. Euphytica 78: 217–226.
Lu Y.H., D’Hont A., Walker D.I.T., Rao P.S., Feldmann P. and Glaszmann J.C. 1994. Relationships among ancestral species of sugarcane revealed with RFLP using single copy maize nuclear probes. Euphytica 78: 7–18.
Martin F.A., Bischoff K.P., Dufrene E.O., Milligan S.B., Quebedeaux J.P., Hoy J.W., Reagan T.E., Giamalva M.J., Miller J.D., Breaux R.D. and Legendre B.L. 1992. Registration of ‘LCP 82-89’ sugarcane. Crop Sci. 32: 499.
McIntyre L. and Jackson P. 1995. Does selfing occur in sugarcane? Plant Genome IV Conference. P165.
Milligan S.B., Martin F.A., Bischoff K.P., Quebedeaux J.P., Dufrene E.O., Quebedeaux K.L., Hoy J.W., Reagan T.E., Legendre B.L. and Miller J.D. 1994. Registration of ‘LCP 85-384’ sugarcane. Crop Sci. 34: 819–820.
Ming R., Liu S.-C., Lin Y.-R., da Silva J., Wilson W., Braga D., van Deynze A., Wenslaff T.F., Wu K.K., Moore P.H., Burnquist W., Sorrels M.E., Irvine J.E. and Paterson A.H. 1998. Detailed alignment ofSaccharum and sorghum chromosomes: comparative organization of closely related diploid and polyploid genomes. Genetics 150: 1663–1682.
Msomi N. and Botha F.C. 1994. Identification of molecular markers linked to fibre using bulk segregant analysis. Proc. S. Afr. Sugar Technol. Assn. 68: 41–45.
Mudge J., Andersen W.R., Kehrer R.L. and Fairbanks D.J. 1996. A RAPD genetic map ofSaccharum officinarum. Crop Sci. 36: 1362–1366.
Nagatomi S. and Ohshiro Y. 1983. Classification of sugarcane wild germplasm by methods of numerical taxonomy. Proc. Intl. Sugar Cane Technol. 18: 650–660.
Pan Y.-B., Burner D.M., Ehrlich K.C., Grisham M.P. and Wei Q. 1997. Analysis of primer-derived, non-specific amplification products in RAPD-PCR. BioTechniques 22: 1071–1077.
Pan Y.-B., Burner D.M. and Legendre B.L. 2000. An assessment of the phylogenetic relationship among sugarcane and related taxa based on the nucleotide sequence of 5S rRNA intergenic spacers. Genetica 108: 285–295.
Pan Y.-B., Cordeiro G., Henry R. and Schnell R.J. 2003. Microsatellite fingerprints of Louisiana sugarcane varieties and breeding lines. Plant and Animal Genome X Conference. W153.
Paran I. and Michelmore R.W. 1993. Development of reliable PCR-based markers linked to downy mildew resistance genes in lettuce. Theor. Appl. Genet. 85: 985–993.
Pillay M. and Kenny S.T. 1995. Anomalies in direct pair-wise comparisons of RAPD fragments for genetic analysis. Biotechniques 19: 694–698.
Rao J.T. and Vijayalakshmi U. 1963. World catalogue of sugarcane genetic stock. Sugarcane Breeding Institute (ICAR), Coimbatore, India, pp. 1–77.
Roxburgh W. 1819. Plants of the Coast of Coromandel, vol. 3, Bulmer, London, pp. 26–27.
Saitou N. and Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425.
Sills G.R., Bridges W., Aljanabi S.M. and Sobral B.W.S. 1995. Genetic analysis of agronomic traits in cross between sugarcane (Saccharum officinarurn L.) and its presumed progenitor (S. robustum Brandes and Jesw. ex Grassl). Mol. Breed. 1: 355–363.
Sobral B.W.S. and Honeycutt R.J. 1993. High output genetic mapping of polyploids using generated markers. Theor. Appl. Genet. 86: 105–112.
Sreenivasan T.V., Ahloowalia B.S. and Heinz D.J. 1987. Cytogenetics. In: Heinz D.J. (ed.), Sugarcane Improvement Through Breeding. Elsevier, New York, pp. 211–253.
Tai P.Y.P., Miller J.D. and Legendre B.L. 1995. Evaluation of the world collection ofSaccharum spontaneum. Proc. Intl. Soc Sugar Cane Technol. 21: 250–260.
Tao Y., Manners J.M., Ludlow M.M. and Henzell R.G. 1993. DNA polymorphisms in grain sorghum (Sorghum bicolor (L.) Moench). Theor. Appl. Genet. 86: 679–688.
Tew T.L. 1987. New varieties. In: Heinz D.J. (ed.), Sugarcane Improvement Through Breeding, Elsevier, New York, pp. 559–594.
Tew T.L. 2004. World sugarcane variety census - Year 2000. Sugar Cane Intl. March/April 2004, pp. 12–18.
Thompson J.D., Higgins D.G. and Gibson T.J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positionspecific gap penalties and weight matrix choice. Nucleic Acid Res. 22(22): 4673–4680.
Author information
Authors and Affiliations
Additional information
Disclaimer: Product names and trademarks are mentioned to report on available data; however, the USDA neither guarantees nor warrants the standard of the product, and the use of the name by USDA does not imply the approval of the product to the exclusion of others that may also be suitable. The experiments reported comply with the current laws of the USA.
Rights and permissions
About this article
Cite this article
Pan, Y.B., Burner, D.M., Legendre, B.L. et al. An assessment of the genetic diversity within a collection ofSaccharum spontaneum L. with RAPD-PCR. Genet Resour Crop Evol 51, 895–903 (2005). https://doi.org/10.1007/s10722-005-1933-1
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s10722-005-1933-1