Abstract
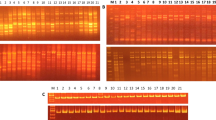
The genetic variability and relationships among 11 cowpea genotypes representing two cultivars and nine elite genotypes were analyzed using 22 random amplified polymorphic DNA (RAPD) and nine inter-simple sequence repeat (ISSR) markers. ISSR markers were more efficient than RAPD assay with regards to polymorphism detection. But the average numbers of polymorphic loci per primer and resolution power were found to be higher for RAPD than for ISSR. Also, the total number of genotype specific marker loci, Nei’s genetic diversity, Shannon’s information index, total heterozygosity, and average heterozygosity were prominent in RAPD as compared to ISSR markers. The regression test between the two Nei’s genetic diversity indices showed low regression (0.3733) between ISSR and RAPD + ISSR-based similarities but maximum (0.9823) for RAPD and RAPD + ISSR-based similarities. The RAPD- and ISSR-generated cultivar- or genotype-specific unique DNA fingerprints able to identify the most diverse genotypes. A dendrogram constructed based on RAPD and ISSR combined data indicated a very clear pattern of clustering according to the groups (cultivars and elite genotypes). The results of principal coordinate analysis were comparable to the cluster analysis. Cluster analysis showed that most diverse genotypes (GP-125 — small size with good seed quality; GP-129, GP-90L — big size with poor seed quality) were separated from moderately diverse cultivars and genotypes. The genetic closeness among GP-129 and GP-90L, JCPL-42, and JCPL-107 could be explained by the high degree of commonness in these genotypes.
Similar content being viewed by others
References
Adawy SS, Hussein Ebtissam HA, El-Khishin Dina Moharam H, El-Itriby Hanayia A. 2002. Genetic variability studies and molecular fingerprinting of some Egyptian date palm (Phoenix dactylifera L.) cultivars: II. RAPD and ISSR profiling. Arab. J. Biotech. 5: 225–236
Ajibade SR, Weeden NF, Chite SM. 2000. Inter simple sequence repeat analysis of genetic relationships in the genus Vigna. Euphytica 111: 47–55
Akkaya MS, Shoemaker RC, Specht JE, Bhagwat AA, Cregan PB. 1995. Integration of simple sequence repeats DNA markers into a soybean linkage map. Crop Sci. 35: 1439–1445
Blair MW, Panaud O, Mccouch SR. 1999. Inter-simple sequence repeat (ISSR) amplification for analysis of microsatellite motif frequency and fingerprinting in rice (Oryza sativa L.). Theor. Appl. Genet. 98: 780–792
Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small amount of fresh leaf tissue. Phytochem. Bull. 19: 11–15
El-Fiky ZA, Hussein Mona H, Mohamed EM, Hussein HA. 2002. Biochemical and molecular genetic studies using SDS-protein, isozymes and RAPD-PCR in some common bean (Phaseolus vulgaris L.) cultivars. Arab. J. Biotech. 5: 249–262
Excoffier L, Smouse PE, Quattro JM. 1992. Analyses of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479–491
Fatokun CA, Danesh D, Young ND, Stewart EL. 1993. Molecular taxonomic relationships in the genus Vigna based on RFLP analysis. Theor. Appl. Genet. 86: 97–104
Galasso I, Pignone D, Perrino P. 1993. Cytotaxonomic studies in Vigna. II. Heterochromatin traitization in Vigna unguiculata and three related species. Carylogia 4: 275–282
Menendez CM, Hall AE, Gepts P. 1997. A genetic linkage map of cowpea (Vigna unguiculata) developed from a cross between 2 inbred, domesticated lines. Theor. Appl. Genet. 95: 1210–1217
Moreno S, Martin JP, Ortiz JM. 1998. Inter-simple sequences repeat PCR for characterization of closely related grapevine germplasm. Euphytica 101: 117–125
Nagaoka T, Ogihara Y. 1997. Applicability of inter-simple sequence repeat polymorphisms in wheat for use as DNA markers in comparison to RFLP and RAPD markers. Theor. Appl. Genet. 94: 597–602
Nei M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individual. Genet. 89: 583–590
Paino D’urzo M, Pedalino M, Grillo S, Rao R, Tucci M. 1990. Variability in major seed proteins in different Vigna species. In NQ Ng, LM Monti, eds, Cowpea Genetic Resources IITA, Ibadan Nigeria, pp 90–100
Panella L, Gepts P. 1992. Genetic relationships within Vigna unguiculata (L.) Walp. based on isozyme analyses. Genet. Resour. Crop. Evol. 39: 71–88
Panella L, Kami J, Gepts P. 1993. Vignin diversity in wild and cultivated taxa of (Vigna unguiculata L.) Walp. (Fabaceae). Econ. Bot. 47: 371–386
Pavlicek A, Heda S, Flegr J. 1999. Free Tree — Freeware program for construction of phylogenetic trees on the basis of distance data and bootstrap/jacknife analysis of tree robustness. Application in the RAPD analysis of genus Frenkelia. Folia Biol.-Praha, 45: 97–99
Peakall R, Smouse PE. 2001. GenAlEx V5: Genetic Analysis in Excel. Population genetic software for teaching and research. Australian National University Canberra, Australia, (http://www.anu.edu.au/BoZo/GenAlEx/)
Prevost A, Wilkinson MJ. 1999. A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor. Appl. Genet. 98: 107–112
Ratnaparkhe MB, Gupta VS, Venmurthy MR, Ranjekar PK. 1995. Genetic fingerprinting of pigeonpea (Cajanus cajan) and its wild relatives using RAPD markers. Theor. Appl. Genet. 91: 893–898
Rohlf FJ (1992). NTSYS-PC: Numerical taxonomy and multivariate analysis system version 2.0. State University of New York Stony Brook, New York
Sharma SK, Dowsons IK, Waugh R. 1995. Relationships among cultivated and wild lentils revealed by RAPD analysis. Theor. Appl. Genet. 91: 647–654
Singh AK, Smart J, Simpson CE, Raina SN. 1998. Genetic variation vis-a-vis molecular polymorphism in groundnut, Arachis hypogaea L. Genet. Resour. Crop Evol. 45: 119–126
Sivaprakash KR, Prasanth SR, Mohanty BP, Parida A. 2004. Genetic diversity of black gram andraces as evaluated by AFLP markers. Curr. Sci. 86: 1411–1415
Sonnante G, Stockton T, Nodari RO, Becerra Velasquez VI, Gepts P. 1994. Evaluation of genetic diversity during the domestication of common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 89: 629–635
Vaillancourt RE, Weeden NF, Barnard J. 1993. Isozyme diversity in the cowpea species complex. Crop Sci. 33: 606–613
Vos R, Hogers R, Bleeker M, Reijans M, Van De Lee T, Hornes M, Fritjers A, Pot J, Peleman J, Kuiper M, Zabeau M. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23: 4407–4414
Wang G, Mahalingam R, Knap HT. 1998. (C-A) and (G-A) anchored simple sequence repeats (ASSRs) generated polymorphism in soybean, Glycine max (L.) Merr. Theor. Appl. Genet. 96: 1086–1096
Williams JGK, Kublick AR, Livak KJ, Rafalski JA, Tingey SV. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18: 6531–6535
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gajera, H.P., Domadiya, R.K., Patel, S.V. et al. Appraisal of RAPD and ISSR markers for genetic diversity analysis among cowpea (Vigna unguiculata L.) genotypes. J. Crop Sci. Biotechnol. 17, 79–88 (2014). https://doi.org/10.1007/s12892-013-0062-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12892-013-0062-1