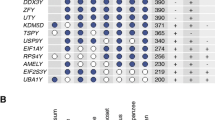
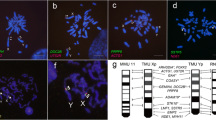
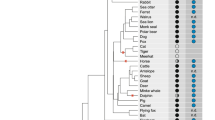
Abstract
Two species of the genus Tokudaia lack the Y chromosome and SRY, but several Y-linked genes have been rescued by translocation or transposition to other chromosomes. Tokudaia muenninki is the only species in the genus that maintains the Y owing to sex chromosome-autosome fusions. According to previous studies, many SRY pseudocopies and other Y-linked genes have evolved by excess duplication in this species. Using RNA-seq and RT-PCR, we found that ZFY, EIF2S3Y, TSPY, UTY, DDX3Y, USP9Y, and RBMY, but not UBA1Y, had high deduced amino acid sequence similarity and similar expression patterns with other rodents, suggesting that these genes were functional. Based on FISH and quantitative real-time PCR, all of the genes except for UTY and DDX3Y were amplified on the X and Y chromosomes with approximately 10–66 copies in the male genome. In a comparative analysis of the 372.4-kb BAC sequence and Y-linked gene transcripts from T. muenninki with the mouse Y genomic sequence, we observed that multiple-copy genes in the ancestral Y genome were nonfunctional, indicating that the gene functions were assumed by amplified copies. We also found a LTR sequence at the distal end of a SRY duplication unit, suggesting that unequal sister chromatid exchange mediated by retrotransposable elements could have been involved in SRY amplification. Our results revealed that the Y-linked genes were rescued from degeneration via translocations to other sex chromosomal regions and amplification events in T. muenninki.






Similar content being viewed by others
Abbreviations
- BAC:
-
Bacterial artificial chromosome
- FISH:
-
Fluorescent in situ hybridization
- LINE:
-
Long interspersed element
- LTR:
-
Long-terminal repeat
- qPCR:
-
Quantitative real-time PCR
- RACE:
-
Rapid amplification of cDNA ends
- RT-PCR:
-
Reverse transcription PCR
- TESCO:
-
The core region within testis-specific enhancer of SOX9
References
Arakawa Y, Nishida-Umehara C, Matsuda Y, Sutou S, Suzuki H (2002) X-chromosomal localization of mammalian Y-linked genes in two XO species of the Ryukyu spiny rat. Cytogenet Genome Res 99:303–309
Bachtrog D (2013) Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet 14:113–124
Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Cho TJ, Koutseva N, Zaghlul S, Graves T, Rock S et al (2014) Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508:494–499
Bianchi NO, Bianchi MS, Bailliet G, de la Chapelle A (1993) Characterization and sequencing of the sex determining region Y gene (Sry) in Akodon (Cricetidae) species with sex reversed females. Chromosoma 102:389–395
Cabot EL, Doshi P, Wu ML, Wu CI (1993) Population genetics of tandem repeats in centromeric heterochromatin: unequal crossing over and chromosomal divergence at the Responder locus of Drosophila melanogaster. Genetics 135:477–487
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K (2009) Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10:421
Chai NN, Zhou H, Hernandez J, Najmabadi H, Bhasin S, Yen PH (1998) Structure and organization of the RBMY genes on the human Y chromosome: transposition and amplification of an ancestral autosomal hnRNPG gene. Genomics 49:283–289
Charlesworth B (2003) The organization and evolution of the human Y chromosome. Genome Biol 4:226
Charlesworth B, Sniegowski P, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371:215–220
Chen HH, Liu TY, Huang CJ, Choo KB (2002) Generation of two homologous and intronless zinc-finger protein genes, zfp352 and zfp353, with different expression patterns by retrotransposition. Genomics 79:18–23
Connallon T, Clark AG (2010) Gene duplication, gene conversion and the evolution of the Y chromosome. Genetics 186:277–286
Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, Grutzner F, Kaessmann H (2014) Origins and functional evolution of Y chromosomes across mammals. Nature 508:488–493
Dobigny G, Ozouf-Costaz C, Bonillo C, Volobouev V (2004) Viability of X-autosome translocations in mammals: an epigenomic hypothesis from a rodent case-study. Chromosoma 113:34–41
Eicher EM, Hutchison KW, Phillips SJ, Tucker PK, Lee BK (1989) A repeated segment on the mouse Y chromosome is composed of retroviral-related, Y-enriched and Y-specific sequences. Genetics 122:181–92
Fernandez R, Barragan MJ, Bullejos M, Marchal JA, Martinez S, Diaz de la Guardia R, Sanchez A (2002) Mapping the SRY gene in Microtus cabrerae: a vole species with multiple SRY copies in males and females. Genome 45:600–603
Fitch DH, Bailey WJ, Tagle DA, Goodman M, Sieu L, Slightom JL (1991) Duplication of the gamma-globin gene mediated by L1 long interspersed repetitive elements in an early ancestor of simian primates. Proc Natl Acad Sci U S A 88:7396–7400
Glusman G, Sosinsky A, Ben-Asher E, Avidan N, Sonkin D, Bahar A, Rosenthal A, Clifton S, Roe B, Ferraz C, Demaille J, Lancet D (2000) Sequence, structure, and evolution of a complete human olfactory receptor gene cluster. Genomics 63:227–45
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q et al (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M et al (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512
Honda T, Suzuki H, Itoh M (1977) An unusual sex chromosome constitution found in the Amami spinous country-rat, Tokudaia osimensis osimensis. Jpn J Genet 52:247–249
Honda T, Suzuki H, Itoh M, Hayashi K (1978) Karyotypical differences of the Amami spinous country-rats, Tokudaia osimensis osimensis obtained from two neighbouring islands. Jpn J Genet 53:297–299
Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Graves T, Fulton RS, Dugan S, Ding Y, Buhay CJ, Kremitzki C (2012) Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature 483:82–86
Hughes JF, Skaletsky H, Koutseva N, Pyntikova T, Page DC (2015) Sex chromosome-to-autosome transposition events counter Y-chromosome gene loss in mammals. Genome Biol 16:104
Just W, Rau W, Vogel W, Akhverdian M, Fredga K, Graves JA, Lyapunova E (1995) Absence of Sry in species of the vole Ellobius. Nat Genet 11:117–118
Kimura R, Murata C, Kuroki Y, Kuroiwa A (2014) Mutations in the testis-specific enhancer of SOX9 in the SRY independent sex-determining mechanism in the genus Tokudaia. PLoS One 9:e108779
Kobayashi T, Yamada F, Hashimoto T, Abe S, Matsuda Y, Kuroiwa A (2008) Centromere repositioning in the X chromosome of XO/XO mammals, Ryukyu spiny rat. Chromosom Res 16:587–593
Kuroiwa A, Ishiguchi Y, Yamada F, Abe S, Matsuda Y (2010) The process of a Y-loss event in an XO/XO mammal, the Ryukyu spiny rat. Chromosoma 119:519–526
Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL (2004) Versatile and open software for comparing large genomes. Genome Biol 5:R12
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 25:1754–60
Marchal JA, Acosta MJ, Bullejos M, Diaz de la Guardia R, Sanchez A (2008) Origin and spread of the SRY gene on the X and Y chromosomes of the rodent Microtus cabrerae: Role of L1 elements. Genomics 91:142–151
Matsuda Y, Chapman VM (1995) Application of fluorescence in situ hybridization in genome analysis of the mouse. Electrophoresis 16:261–272
Matsuda Y, Harada YN, Natsuume-Sakai S, Lee K, Shiomi T, Chapman VM (1992) Location of the mouse compliment factor H gene (cfh) by FISH analysis and replication R-banding. Cell Genet 61:282–285
Murata C, Yamada F, Kawauchi N, Matsuda Y, Kuroiwa A (2010) Multiple copies of SRY on the large Y chromosome of the Okinawa spiny rat, Tokudaia muenninki. Chromosom Res 18:623–634
Murata C, Yamada F, Kawauchi N, Matsuda Y, Kuroiwa A (2012) The Y chromosome of the Okinawa spiny rat, Tokudaia muenninki, was rescued through fusion with an autosome. Chromosom Res 20:111–125
Murata C, Kuroki Y, Imoto I, Tsukahara M, Ikejiri N, Kuroiwa A (2015) Initiation of recombination suppression and PAR formation during the early stages of neo-sex chromosome differentiation in the Okinawa spiny rat, Tokudaia muenninki. BMC Evol Biol 15:234
Noordam MJ, Repping S (2006) The human Y chromosome: a masculine chromosome. Curr Opin Genet Dev 16:225–232
Paria N, Raudsepp T, Pearks Wilkerson AJ, O’Brien PC, Ferguson-Smith MA, Love CC, Arnold C, Rakestraw P, Murphy WJ, Chowdhary BP (2011) A gene catalogue of the euchromatic male-specific region of the horse Y chromosome: comparison with human and other mammals. PLoS One 6:e21374
Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26
Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP et al (2005) The DNA sequence of the human X chromosome. Nature 434:325–337
Rowe KC, Reno ML, Richmond DM, Adkins RM, Steppan SJ (2008) Pliocene colonization and adaptive radiations in Australia and New Guinea (Sahul): multilocus systematics of the old endemic rodents (Muroidea: Murinae). Mol Phylogenet Evol 47:84–101
Rozen S, Skaletsky H, Marszalek JD, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Page DC (2003) Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 423:873–876
Sekido R, Lovell-Badge R (2008) Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 453:930–934
Soullier S, Hanni C, Catzeflis F, Berta P, Laudet V (1998) Male sex determination in the spiny rat Tokudaia osimensis (Rodentia:Muridae) is not Sry dependent. Mamm Genome 9:590–592
Sutou S, Mitsui Y, Tsuchiya K (2001) Sex determination without the Y chromosome in two Japanese rodents Tokudaia osimensis osimensis and Tokudaia osimensis spp. Mamm Genome 12:17–21
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2008) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tsuchiya K, Wakana S, Suzuki H, Hattori S, Hayashi Y (1989) Taxonomic study of Tokudaia (Rodentia: Muridae): I. Genetic differentiation. Memoirs Nat Sci Museum Tokyo 22:227–234
Turner ME, Martin C, Martins AS, Dunmire J, Farkas J, Ely DL, Milsted A (2007) Genomic and expression analysis of multiple Sry loci from a single Rattus norvegicus Y chromosome. BMC Genet 8–11
Verkaar EL, Zijlstra C, van’t Veld EM, Boutaga K, van Boxtel DC, Lenstra JA (2004) Organization and concerted evolution of the ampliconic Y-chromosomal TSPY genes from cattle. Genomics 84:468–474
Vogel W, Jainta S, Rau W, Geerkens C, Baumstark A, Correa-Cerro LS, Ebenhoch C, Just W (1998) Sex determination in Ellobius lutescens: the story of an enigma. Cytogenet Cell Genet 80:214–221
Warburton PE, Waye JS, Willard HF (1993) Nonrandom localization of recombination events in human alpha satellite repeat unit variants: implications for higher-order structural characteristics within centromeric heterochromatin. Mol Cell Biol 13:6520–6529
Yamada F, Kawauchi N, Nakata K, Abe S, Kotaka N, Takashima A, Murata C, Kuroiwa A (2010) Rediscovery after thirty years since the last capture of the critically endangered Okinawa spiny rat Tokudaia muenninki in the northern part of Okinawa Island. Mammal Study 35:243–255
Acknowledgments
The authors thank F. Yamada, T. Jogahara, C. Koshimoto, T. Mitani, and K. Nakata for their efforts in capturing the animals and the Fujii Memorial Institute of Medical Sciences, Tokushima University, for support with fluorescence microscopy. This work was supported by JSPS KAKENHI Grant Numbers 23870023 and 26870410.
Authors’ contributions
CM conceived of and designed the study, performed molecular and cytogenetic experiments and all data analysis, and drafted the manuscript. YK conducted RNA-seq. II participated in total RNA and BAC DNA extraction and commented on the manuscript. AK conceived of and designed the study and participated in drafting the manuscript. All authors read and approved of the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible Editor: Tatsuo Fukagawa, Ph.D
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
Alignment of the nucleotide and amino acid sequences of nine Y-linked genes between T. muenninki and other rodents. The coding region from the mRNA and/or genomic sequences of ZFY (a, b), UBA1Y (c, d), EIF2S3Y (e, f), TSPY (g, h), UTY (i, j), DDX3Y (k, l), USP9Y (m, n), RBMY (o, p), and SRY (q) in T. muenninki were compared with orthologous sequences in mouse, rat, T. osimensis, and/or A. sylvaticus. Identical residues are indicated by dots. TMU: T. muenninki. The deletion and the internal stop codon are colored in gray (a) and pink (m), respectively. Asterisks show the stop codon sites in the amino acid sequences (b, d, f, h, j, l, n, p). Single nucleotide and amino acid polymorphisms within T. muenninki transcripts of each gene are shown in red letters (e–h, m–p). Different nucleotide sites of genomic sequences from the mRNAs of T. muenninki are shown in blue letters (e, g, m, o). (q) The start codons and the stop codons are colored in blue. SRY25 is the genomic sequence observed upstream of EIF2S3Y in this study. SRY1–24 sequences of T. muenninki are reported by Murata et al. 2010, of which only three (SRY1–3) have the conserved coding sequences. The repeat sequence was partially skipped in the mouse (600 bp), and the skipped site is shown by double slashes. (PDF 30457 kb)
Figure S2
Neighbor-joining trees constructed with each X/Y gene sequence of T. muenninki and other rodents. Number above and below branches refer to bootstrap percentages. The values less than 70 % are not shown. TMU T. muenninki; MMU M.musculus; RNO R. norvegicus; ASY A. sylvaticus (GIF 26 kb)
Figure S3
Chromosomal location of two X-linked genes in T. muenninki. A fluorescent in situ hybridization (FISH) analysis showed that ZFX (a, b) and UBA1 (c, d) are distributed on the middle and pericentromeric regions of Xq, respectively. The hybridization signals are indicated by arrowheads. R- (a, c) and G-banding (b, d) patterns are shown. The scale bars represent 10 μm. (GIF 209 kb)
Figure S4
Nucleotide alignment of the accepter splicing site within intron 15 of DDX3Y between T. muenninki and mouse. The intronic sites are colored in gray for each species. Arrows show the boundary between an exon and intron. Asterisks show the identical sites between two sequences. (GIF 29 kb)
Figure S5
Nucleotide sequence alignment in the conserved upstream (a) and downstream (b) regions of SRY among T. muenninki, mouse, and rat. Identical residues are indicated by dots. The skipped site is shown by double slashes. TMU: T. muenninki. (PDF 14453 kb)
Table S1
(XLSX 10 kb)
Table S2
(XLSX 11 kb)
Table S3
(XLSX 10 kb)
Rights and permissions
About this article
Cite this article
Murata, C., Kuroki, Y., Imoto, I. et al. Ancestral Y-linked genes were maintained by translocation to the X and Y chromosomes fused to an autosomal pair in the Okinawa spiny rat Tokudaia muenninki . Chromosome Res 24, 407–419 (2016). https://doi.org/10.1007/s10577-016-9531-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-016-9531-y