Abstract
Key message
An Oligo-FISH barcode system was developed for two model legumes, allowing the identification of all cowpea and common bean chromosomes in a single FISH experiment, and revealing new chromosome rearrangements. The FISH barcode system emerges as an effective tool to understand the chromosome evolution of economically important legumes and their related species.
Abstract
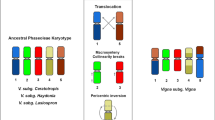
Current status on plant cytogenetic and cytogenomic research has allowed the selection and design of oligo-specific probes to individually identify each chromosome of the karyotype in a target species. Here, we developed the first chromosome identification system for legumes based on oligo-FISH barcode probes. We selected conserved genomic regions between Vigna unguiculata (Vu, cowpea) and Phaseolus vulgaris (Pv, common bean) (diverged ~ 9.7–15 Mya), using cowpea as a reference, to produce a unique barcode pattern for each species. We combined our oligo-FISH barcode pattern with a set of previously developed FISH probes based on BACs and ribosomal DNA sequences. In addition, we integrated our FISH maps with genome sequence data. Based on this integrated analysis, we confirmed two translocation events (involving chromosomes 1, 5, and 8; and chromosomes 2 and 3) between both species. The application of the oligo-based probes allowed us to demonstrate the participation of chromosome 5 in the translocation complex for the first time. Additionally, we detailed a pericentric inversion on chromosome 4 and identified a new paracentric inversion on chromosome 10. We also detected centromere repositioning associated with chromosomes 2, 3, 5, 7, and 9, confirming previous results for chromosomes 2 and 3. This first barcode system for legumes can be applied for karyotyping other Phaseolinae species, especially non-model, orphan crop species lacking genomic assemblies and cytogenetic maps, expanding our understanding of the chromosome evolution and genome organization of this economically important legume group.





Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included as supplementary materials.
References
Albert PS, Zhang T, Semrau K et al (2019) Whole-chromosome paints in maize reveal rearrangements, nuclear domains, and chromosomal relationships. Proc Natl Acad Sci 116:1679–1685. https://doi.org/10.1073/pnas.1813957116
Almeida C, Pedrosa-Harand A (2013) High macro-collinearity between lima bean (Phaseolus lunatus L.) and the common bean (P. vulgaris L.) as revealed by comparative cytogenetic mapping. Theor Appl Genet 126:1909–1916. https://doi.org/10.1007/s00122-013-2106-9
Beliveau BJ, Joyce EF, Apostolopoulos N et al (2012) Versatile design and synthesis platform for visualizing genomes with oligopaint FISH probes. Proc Natl Acad Sci 109:21301–21306. https://doi.org/10.1073/pnas.1213818110
Bi Y, Zhao Q, Yan W et al (2020) Flexible chromosome painting based on multiplex PCR of oligonucleotides and its application for comparative chromosome analyses in Cucumis. Plant J 102:178–186. https://doi.org/10.1111/tpj.14600
Bielski W, Książkiewicz M, Šimoníková D, Hřibová E, Susek K, Naganowska B (2020) The puzzling fate of a lupin chromosome revealed by reciprocal oligo-FISH and BAC-FISH mapping. Genes 11:1489. https://doi.org/10.3390/genes11121489
Bonifácio EM, Fonsêca A, Almeida C et al (2012) Comparative cytogenetic mapping between the lima bean (Phaseolus lunatus L.) and the common bean (P. vulgaris L.). Theor Appl Genet 124:1513–1520. https://doi.org/10.1007/s00122-012-1806-x
Boukar O, Fatokun CA, Huynh B-L et al (2016) Genomic tools in cowpea breeding programs: status and perspectives. Front Plant Sci 7:1–13. https://doi.org/10.3389/fpls.2016.00757
Braz GT, He L, Zhao H et al (2018) Comparative oligo-fish mapping: an efficient and powerful methodology to reveal karyotypic and chromosomal evolution. Genetics 208:513–523. https://doi.org/10.1534/genetics.117.300344
Braz GT, do Vale Martins L, Zhang T et al (2020a) A universal chromosome identification system for maize and wild Zea species. Chromosom Res 28:183–194. https://doi.org/10.1007/s10577-020-09630-5
Braz GT, Yu F, do Vale Martins L, Jiang J (2020b) Fluorescent in situ hybridization using oligonucleotide-based probes. In: Nielsen B, Jones J (eds) In situ hybridization protocols. Methods in molecular biology, vol 2148. Humana, New York, pp 71–83. https://doi.org/10.1007/978-1-0716-0623-0_4
Braz GT, Yu F, Zhao H et al (2021) Preferential meiotic chromosome pairing among homologous chromosomes with cryptic sequence variation in tetraploid maize. New Phytol 229:3294–3302. https://doi.org/10.1111/nph.17098
Broughton WJ, Hern A et al (2003) Beans (Phaseolus spp.): model food legumes. Plant Soil 252:55–128
Chen L, Su D, Sun J et al (2020) Development of a set of chromosome-specific oligonucleotide markers and karyotype analysis in the Japanese morning glory Ipomoea nil. Sci Hortic (amsterdam) 273:109633. https://doi.org/10.1016/j.scienta.2020.109633
Darlington CD, Wylie AP (1955) Chromosome atlas of cultivated plants, 2nd edn. Chromosome atlas of flowering plants, London
De Carvalho CR, Saraiva LS (1993) An air drying technique for maize chromosomes without enzymatic maceration. Biotech Histochem 68:142–145. https://doi.org/10.3109/10520299309104684
do Vale Martins L, Yu F, Zhao H et al (2019) Meiotic crossovers characterized by haplotype-specific chromosome painting in maize. Nat Commun 10:4604. https://doi.org/10.1038/s41467-019-12646-z
do Vale Martins L, Bustamante FO, Oliveira ARS, et al (2021) BAC- and oligo-FISH mapping reveals chromosome evolution among Vigna angularis, V. unguiculata and Phaseolus vulgaris. Chromosoma. https://doi.org/10.1007/s00412-021-00758-9
Ferraz ME, Fonsêca A, Pedrosa-Harand A (2020) Multiple and independent rearrangements revealed by comparative cytogenetic mapping in the dysploid Leptostachyus group (Phaseolus L., Leguminosae). Chromosom Res 28:395–405. https://doi.org/10.1007/s10577-020-09644-z
Fonsêca A, Ferraz ME, Pedrosa-Harand A (2016) Speeding up chromosome evolution in Phaseolus: multiple rearrangements associated with a one-step descending dysploidy. Chromosoma 125:413–421. https://doi.org/10.1007/s00412-015-0548-3
Fonsêca A, Ferreira J, dos Santos TRB et al (2010) Cytogenetic map of common bean (Phaseolus vulgaris L.). Chromosom Res 18:487–502. https://doi.org/10.1007/s10577-010-9129-8
Fonsêca A, Pedrosa-Harand A (2013) Karyotype stability in the genus Phaseolus evidenced by the comparative mapping of the wild species Phaseolus microcarpus. Genome 56:335–343. https://doi.org/10.1139/gen-2013-0025
Forni-Martins ER (1986) New chromosome number in the genus Vigna Savi (Leguminosae-Papilionoideae). Bull Du Jard Bot Natl Belgique/Bull Van Natl Plantentuin Van België 56:129. https://doi.org/10.2307/3667759
Filho FRF, Ribeiro VQ, de Rocha MM et al (2011) Feijão-Caupi no Brasil, 1st edn. EMBRAPA Meio-Norte, Teresina
Garcia T, Duitama J, Zullo S et al (2020) Comprehensive genomic resources related to domestication and crop improvement traits in Lima bean. Nat Res. https://doi.org/10.1038/s41467-021-20921-1
Geffroy V, Macadré C, David P et al (2009) Molecular analysis of a large subtelomeric nucleotide-binding-site–leucine-rich-repeat family in two representative genotypes of the major gene pools of Phaseolus vulgaris. Genetics 181:405–419. https://doi.org/10.1534/genetics.108.093583
Gepts P, Aragão FJL, de Barros E et al (2008) Genomics of Phaseolus beans, a major source of dietary protein and micronutrients in the tropics. In: Moore PH, Ming R (eds) Genomics of tropical crop plants. Springer, New York, pp 113–143
Han Y, Zhang T, Thammapichai P et al (2015) Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics 200:771–779. https://doi.org/10.1534/genetics.115.177642
Hasing T, Tang H, Brym M et al (2020) A phased Vanilla planifolia genome enables genetic improvement of flavour and production. Nat Food 1:811–819. https://doi.org/10.1038/s43016-020-00197-2
He L, Braz GT, Torres GA, Jiang J (2018) Chromosome painting in meiosis reveals pairing of specific chromosomes in polyploid Solanum species. Chromosoma 127:505–513. https://doi.org/10.1007/s00412-018-0682-9
He L, Zhao H, He J et al (2020) Extraordinarily conserved chromosomal synteny of Citrus species revealed by chromosome-specific painting. Plant J 103:2225–2235. https://doi.org/10.1111/tpj.14894
Hou L, Xu M, Zhang T et al (2018) Chromosome painting and its applications in cultivated and wild rice. BMC Plant Biol 18:110. https://doi.org/10.1186/s12870-018-1325-2
Hougaard BK, Madsen LH, Sandal N et al (2008) Legume anchor markers link syntenic regions between Phaseolus vulgaris, Lotus japonicus, Medicago truncatula and Arachis. Genetics 179:2299–2312. https://doi.org/10.1534/genetics.108.090084
Iwata-Otsubo A, Radke B, Findley S et al (2016) Fluorescence in situ hybridization (FISH)-based karyotyping reveals rapid evolution of centromeric and subtelomeric repeats in common bean (Phaseolus vulgaris) and relatives. G3 Genes Gen Genet 6:1013–1022. https://doi.org/10.1534/g3.115.024984
Jiang J (2019) Fluorescence in situ hybridization in plants: recent developments and future applications. Chromosom Res 27:153–165. https://doi.org/10.1007/s10577-019-09607-z
Jiang J, Gill BS (1994) Nonisotopic in situ hybridization and plant genome mapping: the first 10 years. Genome 37:717–725. https://doi.org/10.1139/g94-102
Jiang J, Gill BS (2006) Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 49:1057–1068. https://doi.org/10.1139/g06-076
Kirov I, Khrustaleva L, Van Laere K et al (2017) DRAWID: user-friendly java software for chromosome measurements and idiogram drawing. Comp Cytogenet 11:747–757. https://doi.org/10.3897/compcytogen.v11i4.20830
Krzywinski M, Schein J, Birol I et al (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. https://doi.org/10.1101/gr.092759.109
Langer-Safer PR, Levine M, Ward DC (1982) Immunological method for mapping genes on Drosophila polytene chromosomes. Proc Natl Acad Sci 79:4381–4385. https://doi.org/10.1073/pnas.79.14.4381
Li H, Wang W, Lin L et al (2013) Diversification of the phaseoloid legumes: effects of climate change, range expansion and habit shift. Front Plant Sci 4:1–9. https://doi.org/10.3389/fpls.2013.00386
Li G, Zhang T, Yu Z et al (2020) An efficient Oligo-FISH painting system for revealing chromosome rearrangements and polyploidization in Triticeae. Plant J 105:978–993. https://doi.org/10.1111/tpj.15081
Liu X, Sun S, Wu Y et al (2020) Dual-color oligo-FISH can reveal chromosomal variations and evolution in Oryza species. Plant J 101:112–121. https://doi.org/10.1111/tpj.14522
Lonardi S, Muñoz-Amatriaín M, Liang Q et al (2019) The genome of cowpea ( Vigna unguiculata [L.] Walp.). Plant J 98:767–782. https://doi.org/10.1111/tpj.14349
Mandáková T, Hloušková P, Koch MA, Lysak MA (2020) Genome evolution in Arabideae was marked by frequent centromere repositioning. Plant Cell 32:650–665. https://doi.org/10.1105/tpc.19.00557
Meng Z, Zhang Z, Yan T et al (2018) Comprehensively characterizing the cytological features of Saccharum spontaneum by the development of a complete set of chromosome-specific oligo probes. Front Plant Sci 9:1–11. https://doi.org/10.3389/fpls.2018.01624
Meng Z, Han J, Lin Y et al (2020) Characterization of a Saccharum spontaneum with a basic chromosome number of x = 10 provides new insights on genome evolution in genus Saccharum. Theor Appl Genet 133:187–199. https://doi.org/10.1007/s00122-019-03450-w
Mercado-Ruaro P, Delgado-Salinas A (1996) Karyological studies in several Mexican species of Phaseolus L. and Vigna Savi (Phaseolinae, Fabaceae). In: Pickergill B, Lock JM (eds) Advances in legume systematics 8, Legumes of economic importance. Royal Botanic Gardens, Kew, pp 83–87
Mercado-Ruaro P, Delgado-Salinas A (1998) Karyotypic studies on species of Phaseolus (Fabaceae: Phaseolinae). Am J Bot 85:1–9. https://doi.org/10.2307/2446547
Mercado-Ruaro P, Delgado-Salinas A (2000) Cytogenetic studies in Phaseolus L. (Fabaceae). Genet Mol Biol 23:985–987. https://doi.org/10.1590/S1415-47572000000400043
Murray J, Larsen J, Michaels TE et al (2002) Identification of putative genes in bean ( Phaseolus vulgaris ) genomic (Bng) RFLP clones and their conversion to STSs. Genome 45:1013–1024. https://doi.org/10.1139/g02-069
Oliveira ARS, do Vale Martins LV, Bustamante FDO et al (2020) Breaks of macrosynteny and collinearity among moth bean (Vigna aconitifolia), cowpea (V. unguiculata), and common bean (Phaseolus vulgaris). Chromosom Res. https://doi.org/10.1007/s10577-020-09635-0
Pecrix Y, Staton SE, Sallet E et al (2018) Whole-genome landscape of Medicago truncatula symbiotic genes. Nat Plants 4:1017–1025. https://doi.org/10.1038/s41477-018-0286-7
Piperidis N, D’Hont A (2020) Sugarcane genome architecture decrypted with chromosome-specific oligo probes. Plant J 103:2039–2051. https://doi.org/10.1111/tpj.14881
Qin S, Wu L, Wei K et al (2019) A draft genome for Spatholobus suberectus. Sci Data 6:1–9. https://doi.org/10.1038/s41597-019-0110-x
Qu M, Li K, Han Y et al (2017) Integrated karyotyping of woodland strawberry (Fragaria vesca) with oligopaint FISH probes. Cytogenet Genome Res 153:158–164. https://doi.org/10.1159/000485283
Schneider KL, Xie Z, Wolfgruber TK, Presting GG (2016) Inbreeding drives maize centromere evolution. Proc Natl Acad Sci 113:E987–E996. https://doi.org/10.1073/pnas.1522008113
Schmutz J, McClean PE, Mamidi S et al (2014) A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet 46:707–713. https://doi.org/10.1038/ng.3008
Schubert I (2018) What is behind “centromere repositioning”? Chromosoma 127:229–234. https://doi.org/10.1007/s00412-018-0672-y
Schubert I, Lysak MA (2011) Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet 27:207–216
Šimoníková D, Němečková A, Karafiátová M et al (2019) Chromosome painting facilitates anchoring reference genome sequence to chromosomes in situ and integrated karyotyping in banana (Musa spp.). Front Plant Sci 10:1–12. https://doi.org/10.3389/fpls.2019.01503
Song X, Song R, Zhou J et al (2020) Development and application of oligonucleotide-based chromosome painting for chromosome 4D of Triticum aestivum L. Chromosom Res 28:171–182. https://doi.org/10.1007/s10577-020-09627-0
Talbert PB, Henikoff S (2020) What makes a centromere? Exp Cell Res 389:111895. https://doi.org/10.1016/j.yexcr.2020.111895
Vallejos CE, Sakiyama NS, Chase CD (1992) A molecular marker-based linkage map of Phaseolus vulgaris L. Genetics 131:733–740
Varshney RK, Chen W, Li Y et al (2012) Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat Biotechnol 30:83–89. https://doi.org/10.1038/nbt.2022
Vasconcelos EV, de Andrade Fonsêca AF, Pedrosa-Harand A et al (2015) Intra- and interchromosomal rearrangements between cowpea [Vigna unguiculata (L.) Walp.] and common bean (Phaseolus vulgaris L.) revealed by BAC-FISH. Chromosom Res 23:253–266. https://doi.org/10.1007/s10577-014-9464-2
Venora G, Blangifortil S, Cremonini R (1999) Karyotype analysis of twelve species belonging to genus Vigna. Cytologia (tokyo) 64:117–127. https://doi.org/10.1508/cytologia.64.117
Wang J, Roe B, Macmil S et al (2010) Microcollinearity between autopolyploid sugarcane and diploid sorghum genomes. BMC Genomics 11:261. https://doi.org/10.1186/1471-2164-11-261
Wu F, Tanksley SD (2010) Chromosomal evolution in the plant family Solanaceae. BMC Genomics 11:182. https://doi.org/10.1186/1471-2164-11-182
Xin H, Zhang T, Han Y et al (2018) Chromosome painting and comparative physical mapping of the sex chromosomes in Populus tomentosa and Populus deltoides. Chromosoma 127:313–321. https://doi.org/10.1007/s00412-018-0664-y
Yang L, Koo D-H, Li D et al (2014) Next-generation sequencing, FISH mapping and synteny-based modeling reveal mechanisms of decreasing dysploidy in Cucumis. Plant J 77:16–30. https://doi.org/10.1111/tpj.12355
Zhao Q, Wang Y, Bi Y et al (2019) Oligo-painting and GISH reveal meiotic chromosome biases and increased meiotic stability in synthetic allotetraploid Cucumis ×hytivus with dysploid parental karyotypes. BMC Plant Biol 19:471. https://doi.org/10.1186/s12870-019-2060-z
Acknowledgements
We thank Embrapa Meio-Norte (Teresina, Piauí, Brazil) and Embrapa Cenargen (Brasília, Distrito Federal, Brazil) for providing the V. unguiculata and P. vulgaris seeds, respectively. We thank CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) Grant No. 421968/2018-4, 313527/2017-2, 310804/2017-5, 433931/2018-3, and 442019/2019-0, and FACEPE (Fundação de Amparo à Ciência e Tecnologia do Estado de Pernambuco) Grant No. APQ-0390-2.02/19 for the financial support. We also thank CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Financial Code 001), and FACEPE for the scholarships.
Funding
This work was supported by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) Grant No. 421968/2018-4, 313527/2017-2, 310804/2017-5, 433931/2018-3, and 442019/2019-0, and FACEPE (Fundação de Amparo à Ciência e Tecnologia do Estado de Pernambuco) Grant No. APQ-0390-2.02/19.
Author information
Authors and Affiliations
Contributions
FOB: designed oligo system, provided resources for the oligo-FISH experiments and wrote the paper. THN: conducted oligo-FISH experiments, analyzed the sequence synteny data, constructed Circos images and wrote the paper. CCMJ: analyzed the sequence synteny data, constructed the Circos images and helped writing the paper. SD: conducted the oligo-FISH experiments and analyzed the sequence synteny data. LVM: analyzed and processed the oligo-FISH images and helped writing the paper. GTB: labeled the oligo-FISH probes. AMBI: discussed the results. JJ: discussed the results and provided resources for the oligo probe labeling. APH: designed this research, wrote the manuscript, discussed the results and provided resources for the oligo-FISH experiments. ACBV: designed this research, the images, wrote the manuscript and discussed the results. All authors read and approved the final version of the paper.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Communicated by P. Heslop-Harrison.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
122_2021_3921_MOESM1_ESM.xlsx
Genomic positions of oligo barcode and BAC markers in the reference genome of Vigna unguiculata (Vu) showing the scores of BLAST search, as well as the positions in bp. a Total size and centromere regions by Lonardi et al. (2019). b BACs from V. unguiculata were previous used by Oliveira et al. (2020) and do Vale Martins et al. (2021) and are available at HarvEST:cowpea (harvest.ucr.edu). The color of each chromosome marker was defined in accordance with Fig. 2 description. (XLSX 21 KB)
122_2021_3921_MOESM2_ESM.xlsx
Genomic positions of oligo barcode and BAC markers in the reference genome of Phaseolus vulgaris (Pv) showing the scores of BLAST search, as well as the positions in bp. Genomic positions of oligo barcode and BAC markers in the reference genome of Phaseolus vulgaris (Pv) showing the scores of BLAST search, as well as the positions in bp. a Total size and centromere regions by Schmutz et al. (2014). c Gene markers ID and accessions for BAC sequences available at NCBI GenBank and EMBL-EBI. The color of each chromosome marker was defined in accordance with Fig. 2 description. (XLSX 21 KB)
122_2021_3921_MOESM3_ESM.xlsx
Chromosome positions of oligo barcode and BAC markers on Vigna unguiculata idiogram. Total chromosome size and position of centromere in bp are also indicated. The color of each chromosome marker was defined in accordance with Fig. 2 description. (XLSX 19 KB)
122_2021_3921_MOESM4_ESM.xlsx
Chromosome positions of oligo barcode and BAC markers on Phaseolus vulgaris idiogram. Total chromosome size and position of centromere in bp are also indicated. The color of each chromosome marker was defined in accordance with Fig. 2 description. (XLSX 19 KB)
122_2021_3921_MOESM5_ESM.pdf
Sequences of the green oligo set used for oligo-FISH of V. unguiculata and P. vulgaris chromosomes. The sequences were selected using the reference genome of V. unguiculata ‘IT97K-499-35’ (Lonardi et al. 2019), based on regions conserved in P. vulgaris (Schmutz et al. 2014) (PDF 8332 KB)
122_2021_3921_MOESM6_ESM.pdf
Sequences of the red oligo set used for oligo-FISH of V. unguiculata and P. vulgaris chromosomes. The sequences were selected using the reference genome of V. unguiculata ‘IT97K-499-35’ (Lonardi et al. 2019), based on regions conserved in P. vulgaris (Schmutz et al. 2014) (PDF 8148 KB)
Rights and permissions
About this article
Cite this article
de Oliveira Bustamante, F., do Nascimento, T.H., Montenegro, C. et al. Oligo-FISH barcode in beans: a new chromosome identification system. Theor Appl Genet 134, 3675–3686 (2021). https://doi.org/10.1007/s00122-021-03921-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-021-03921-z



