Abstract
The inactivation of one of the two X chromosomes in female placental mammals represents a remarkable example of epigenetic silencing. X inactivation occurs also in marsupial mammals, but is phenotypically different, being incomplete, tissue-specific and paternal. Paternal X inactivation occurs also in the extraembryonic cells of rodents, suggesting that imprinted X inactivation represents a simpler ancestral mechanism. This evolved into a complex and random process in placental mammals under the control of the XIST gene, involving notably variant and modified histones. Molecular mechanisms of X inactivation in marsupials are poorly known, but occur in the absence of an XIST homologue. We analysed the specific pattern of histone modifications using immunofluorescence on metaphasic chromosomes of a model kangaroo, the tammar wallaby. We found that all active marks are excluded from the inactive X in marsupials, as in placental mammals, so this represents a common feature of X inactivation throughout mammals. However, we were unable to demonstrate the accumulation of inactive histone marks, suggesting some fundamental differences in the molecular mechanism of X inactivation between marsupial and placental mammals. A better understanding of the epigenetic mechanisms underlying X inactivation in marsupials will provide important insights into the evolution of this complex process.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
X chromosome inactivation is an important model system for studying epigenetic repression. In female mammals, one of the two X chromosomes is silenced, achieving dosage compensation of X-borne genes between females (XX) and males (XY) (Lyon 1961; reviewed by Heard and Disteche 2006). Silencing is at the level of transcription (Graves and Gartler 1986). Once initiated, the transcriptional silencing of thousands of X-linked genes is heritably maintained through all subsequent cell divisions. The molecular mechanism of this developmental regulation in mouse and human females is now known to involve histone variation and modification. How these modifications achieve transcriptional silencing is of great interest, as is how such a complex regulatory system evolved in mammals.
The developmentally regulated (facultative) heterochromatin state of the inactive X is brought about by a complex of molecular changes to chromatin. During the past decade, increasing numbers of studies have shown that histone modifications, such as acetylation, methylation, phosphorylation and ubiquitinylation, play a key role in all DNA-based processes, directly via the disruption of chromatin contacts, or indirectly via the recruitment of effector proteins (reviewed by Nightingale et al. 2006; Kouzarides 2007). Notably, fine combinations of these different modifications participate largely in the regulation of transcription, at a local or global scale (reviewed by Berger 2007). Indeed, they lead to activation or repression of specific genes or entire genomic regions (called respectively euchromatin or heterochromatin).
Histone modifications have been found to have both short-term and long-term effects on gene function (reviewed by Turner 2007). Long-term effects define and maintain stable chromatin structures throughout the cell cycle. Indeed, histone modifications can be efficiently stable and trigger the memory of transcription profiles through cell divisions. Thus, they can act as powerful epigenetic marks that lead to stable and heritable changes of gene expression without alteration of gene sequence. Dissection of these epigenetic mechanisms is essential for our understanding of complex gene expression pathways underlying the extraordinary diversity of cell lineages that form a multicellular organism.
Nearly all the studies of the molecular mechanism of X chromosome inactivation (XCI) have been carried out on mice and humans. In these and other placental mammals, XCI occurs during early embryonic development and can be recapitulated during differentiation of embryonic stem (ES) cells (Okamoto et al. 2004; Rastan and Robertson 1985). Initiation and propagation of inactivation is controlled in cis by a complex locus (the X inactivation centre, XIC). This locus contains the critical gene XIST, which produces a noncoding RNA responsible for triggering the silencing signal (Brown et al. 1991; Chureau et al. 2002). The first observable event is the accumulation of XIST RNA along the X chromosome to be inactivated, and this is closely followed by several changes in histone variants and modifications (reviewed by Heard 2005). First, histone modifications associated with transcriptional activity (H3K4me2, H3K9ac) are lost (Heard et al. 2001; Chaumeil et al. 2002; Okamoto et al. 2004). Soon after, the inactive X gains specific repressive marks, H3K9me2/H3K27me3/H4K20me1/H2AK119ub (Heard et al. 2001; Mermoud et al. 2002; Plath et al. 2003; Silva et al. 2003; Okamoto et al. 2004; Rougeulle et al. 2004; Kohlmaier et al. 2004; de Napoles et al. 2004; Fang et al. 2004; Hernandez-Munoz et al. 2005). Finally, it loses acetylation of histone H4 (H4Kac), which is associated with active chromatin (Keohane et al. 1996; Heard et al. 2001; Chaumeil et al. 2002; Okamoto et al. 2004). The role of histone modifications in the inactivation process is not yet understood, but it may confer the stabilization that underpins the somatic heritability of the inactive state (Kohlmaier et al. 2004). Finally, other changes appear, including a shift to late replication timing (Keohane et al. 1996; Chaumeil et al. 2002; Okamoto et al. 2004), recruitment of the histone variant macroH2A (Costanzi and Pehrson 1998; Mermoud et al. 1999), and CpG island methylation of DNA (Lock et al. 1987; Norris et al. 1991; Sado et al. 2004).
Novel insight into the mechanism of X inactivation can be gained by comparing its components across larger evolutionary distances. XCI occurs in marsupials as well as placental mammals, but is absent outside these therian mammals. In the egg-laying monotremes, basal mammals that diverged from therians 166 Mya, multiple X chromosomes have homology not to the human X, but to the chicken Z chromosome (Veyrunes et al. 2008). Like the chicken Z (Itoh et al. 2007), platypus X chromosomes do not undergo inactivation, although some form of partial dosage compensation operates (Deakin et al. 2008).
Marsupial XCI shows many phenotypic and molecular differences from human and mouse XCI, so offers insights into how XCI works and how it evolved. As for humans and mice, the marsupial inactive X replicates late in S phase (Graves 1967; Sharman 1971) and sex chromatin has been observed, though inconsistently, in some species and tissues (VandeBerg et al. 1987). Differential histone H4 underacetylation has also been demonstrated between the two X chromosomes in cells from female marsupials (Wakefield et al. 1997), but no DNA methylation differences have been detected (Piper et al. 1993; Loebel and Johnston 1996). Phenotypically, marsupial XCI differs fundamentally from placental XCI in being incomplete and tissue-specific (reviewed by Cooper et al. 1993), and less stable (Kaslow and Migeon 1987).
Perhaps the most dramatic difference is that marsupial XCI is imprinted, as it is the paternal X chromosome that is always the inactivate one (Cooper 1971; Sharman 1971), whereas inactivation is random in placental mammals. The subsequent discovery that paternal X inactivation also occurred during the first stages of rodent development and was retained in rodent extraembryonic tissues (Takagi and Sasaki 1975; Huynh and Lee 2003; Okamoto et al. 2004) suggests imprinted XCI in marsupials may represent a simpler ancestral inactivation mechanism that evolved into a more complex and random process in placental embryos. It was proposed (Cooper 1971) that the marsupial paternal X, as well as the Y, is inactivated during male meiosis and stays inactive throughout fertilization and development. This link has been explored during early mouse development, but yielded inconsistent results. One study showed that the paternal X arrives already inactive in the zygote (Huynh and Lee 2003), but another showed that the paternal X is active at the 2-cell stage and that initiation of its inactivation at the 4-cell stage is independent of its inactivation during meiosis (Okamoto et al. 2004, 2005).
A major difference between inactivation mechanisms in placental and marsupial mammals is the apparent absence of the XIST gene from the marsupial X chromosome, recently confirmed by three independent studies. Physical mapping and bioinformatic analyses revealed that the region homologous to the X inactivation centre is disrupted by independent rearrangements in marsupials and monotremes (Hore et al. 2007; Shevchenko et al. 2007), and it was suggested that XIST evolved from a protein coding gene (Lnx3) in this region, whose loss of function coincided with pseudogenization of flanking genes (Duret et al. 2006). These findings challenge a pivotal role in X inactivation for XIST and raise many fundamental questions about the mechanism and evolution of this important epigenetic system. Firstly, without a marsupial XIST homologue, how is XCI initiated in marsupials? And how, without XIST RNA, are variant and modified histones recruited?
As part of an ongoing molecular characterization of marsupial XCI, we determined the binding patterns of several key histone modifications on marsupial metaphase chromosomes, in order to assess their involvement in marsupial XCI and the relationship of these changes to XIST and DNA methylation.
Materials and methods
Cell culture
We focused on the tammar wallaby Macropus eugenii, the model kangaroo (family Macropodidae) whose genome is being sequenced. Fibroblasts cells were obtained from ear tissue of males and females under permit WT20006-6627. Cells were cultured at 35°C in 5% CO2 in a 1:1 mix of Dulbecco’s Modified Eagle’s (DME) and Amniomax C100 media (Gibco Invitrogen, Carlsbad, CA, USA), supplemented with 10% fetal calf serum (FCS; Gibco Invitrogen).
Immunofluorescence on cytospin prepared metaphase
Chromosome preparations
Tammar wallaby fibroblast cells (1× T75 flask) were treated with Colcemid (final concentration 0.1 mg/ml) for 1 h at 35°C. Cells were gently harvested and pelleted for 5 min at 400g. The pellet was resuspended in a few drops of residual medium and placed on ice until 3 ml of ice cold 75 mM KCl was added in a drop-wise manner with gentle agitation. Cells were allowed to swell at 37°C for 15 min and then placed on ice. Cells were diluted to a density of 1 × 105 cells/ml in 75 mM KCl/0.1% Tween 20 before loading 250 μl into a cytofunnel, and centrifuged at 800 rpm for 3 min in a Cytospin 3 Cell Preparation System centrifuge (Thermo Shandon, Waltham, MA, USA).
Immunofluorescence
Immunofluorescences on metaphase spreads were performed as previously described (Chaumeil et al. 2002, 2004) with minor modifications. Slides were allowed to dry for 5 min before transferring them to KCM pH 7.5 (10 mM Tris HCl pH 7.6, 120 mM KCl, 20 mM NaCl, 0.5 mM EDTA, 0.1% Triton) for 15 min at RT. Slides were blotted around cell area before incubating with primary antibodies (Upstate-Millipore, Billerica, MA, USA) diluted 1/200 (1/400 for H3K4me2) in KCM/ 10% FCS. Primary antibody was incubated for 2 h at 37°C in a humid chamber (antibodies are listed in Table 1). Slides were washed twice in KCM for 5 min each at RT before incubating with secondary antibody, anti-rabbit Texas red (Jackson Immunoresearch Laboratories, West Grove, PA, USA) for 30 min at RT. Slides were washed twice in KCM for 5 min each and then fixed in 4% paraformaldehyde/KCM for 15 min before they were rinsed in distilled water, air dried and mounted in DAPI (1.5 μg/ml) containing Vectashield (Vector Laboratories, Burlingame, CA, USA).
Cytogenetic analysis
Fluorescence was visualized with a Ziess Axioplan epifluorescence microscope and images recorded with a thermoelectronically-cooled charge-coupled device camera (RT Monochrome Spot, Diagnostic Instruments, Sterling Heights, MI, USA). IPLab imaging software (Scanalytics Inc. Fairfax, VA, USA) was used to capture grey-scale images and to convert them to coloured images that could be superimposed. Line scans of DAPI and histone modifications intensities were designed using Softworx software (Applied Precision, Issaquah, WA, USA).
All antibodies were hybridized to both female and male-derived metaphases. In each hybridization experiment female and male-derived cells were placed on the same slide for consistency of reagents and conditions. Any differences seen in hybridizations between the two X chromosomes in female-derived metaphases were compared back to the single active X chromosome of male-derived cells.
Three independent experiments were performed for each antibody. Twenty metaphases were counted for each experiment.
Results
The tammar wallaby (2 n = 16) has seven pairs of autosomes and one pair of sex chromosomes. The X chromosome is the only small submetacentric chromosome, and is easily distinguished by its morphology and the prominent interstitial nucleolus organizer region (NOR) on its short arm (Alsop et al. 2005). The C- band positive short arm of the X (Xp) consists of sequences shared with the Y chromosome, which were added relatively recently to both sex chromosomes (Toder et al. 1997). The long arm of the X chromosome (Xq) contains the suite of conserved genes shared with human Xq. The two X chromosomes in female tammar cells replicate asynchronously (Graves 1967). Studies of Xq genes in a variety of macropodid marsupials demonstrated that different genes on the X are inactive in different tissues, whereas the NOR on Xp is active on both chromosomes (VandeBerg et al. 1987; Graves and Dawson 1988; Cooper et al. 1993; Koina et al. 2005).
Profile of active histone marks on the marsupial inactive X chromosome
In humans and mice, the inactive X is depleted in histone H3 dimethylated at lysine 4 (H3K4me2), as well as in acetylated H3 and H4, three histone modifications that are associated with active chromatin (Jeppesen and Turner 1993; Heard et al. 2001; Boggs et al. 2002; Chaumeil et al. 2002; Okamoto et al. 2004). We performed immunofluorescences on metaphase spreads in order to investigate their status on the marsupial inactive X chromosome (n = 20 for each of the 3 independent experiments done).
Histone H3 dimethylation at lysine 4 (H3K4me2)
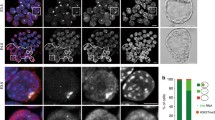
The antibody directed against H3K4me2 hybridized in very consistent and specific patterns on the arms, but not the centromeres, of all autosomal pairs (Fig. 1A).This antibody revealed strikingly differential binding profiles between the two X chromosomes in tammar female metaphase spreads (Fig. 1Ad). One of the X chromosomes showed strong staining over Xq (particularly the distal region and a narrow proximal region) but none on Xp (>90% of metaphases). Similarity of this histone methylation pattern to that of the single X chromosome in male-derived cells (Fig. 1Ah) implies that this stained chromosome is the active X. The other X showed no staining, implying that the inactive X is depleted in H3K4me2. Similarly, the Y chromosome of males is depleted in H3K4me2. Thus the active histone mark H3K4me2 is restricted to the active X chromosome.
Status of active marks on the inactive X chromosome in tammar wallaby. A Histone H3 dimethylated on lysine 4 (H3K4me2). B Histone H4 acetylated on lysines (H4Kac). C Histone H3 acetylated on lysine 9 (H3K9ac). In each case, the top row shows metaphase spread from female cells and the bottom row shows metaphase spread from male cells. a and e DAPI staining of chromosomes; b and f antibody staining; c and g merged picture (DAPI in red, immunofluorescence in green); d enlarged view of X chromosomes in females with line scans for DAPI and histone modification intensities (Xa=active X, Xi=inactive X); h enlarged view of X (Xa=active X) and Y chromosomes in males with line scans for DAPI and histone modification intensities. * shows hybridization hotspots. Scale bar represents 10 μm
Histone H4 acetylation (H4Kac)
Previously it was shown that the tammar inactive X is underacetylated on H4 (Wakefield et al. 1997). We included the same antibody directed against H4 lysine acetylation (H4Kac) in our experimental plan as a control for our hybridization conditions. We observed complex and consistent binding patterns on all tammar autosome arms, but no binding on centromeres or on the Y chromosome (Fig. 1B). The two X chromosomes in female cells showed quite different hybridization patterns (>90% of metaphases). One X chromosome showed staining on Xq (particularly the distal region and a thin proximal band, similar to the pattern produced by H3K4me2) but not on Xp or the NOR (Fig. 1Bd). This acetylation pattern corresponded to that observed on the single active X in males (Fig. 1Bh). In females, the other X showed no staining at all, confirming that the inactive X is depleted of H4Kac.
Histone H3 acetylation (H3K9ac)
The antibody directed against H3 acetylation at lysine 9 (H3K9ac) also showed distinct and consistent patterns on the arms of autosomal pairs, with no centromeric staining (Fig. 1C). In females the two X chromosomes had different binding patterns (>90% of metaphases). One X chromosome was stained over the entire Xq arm, with no staining over Xp (Fig. 1Cd). As this pattern was similar to the active X chromosome in male-derived cells, it must correspond to the active X chromosome. In contrast, the staining on the other X chromosome was restricted to one hotspot below the centromere and a second on the distal end of Xq. There was no binding on Xp or the Y chromosome (Fig. 1Ch). Thus acetylated histone H3 is absent from the inactive X.
In summary then, all the antibodies recognizing active histone marks were excluded from the inactive X chromosome of the female tammar wallaby.
Profile of inactive histone marks on the marsupial inactive X chromosome
To determine whether modifications associated with silent chromatin accumulated on the tammar inactive X chromosome, as they do on the human and mouse inactive X, we tested four antibodies against methylated histones on metaphase spreads (n = 20 for each of the 3 independent experiments done). These antibodies included two modifications known to be enriched on the inactive X of placental mammals, H3 dimethylation of lysine 9 (H3K9me2) and H3 trimethylation of lysine 27 (H3K27me3) (Heard et al. 2001; Mermoud et al. 2002; Plath et al. 2003; Silva et al. 2003; Okamoto et al. 2004; Rougeulle et al. 2004) and two modifications known to be specific to the pericentric heterochromatin, H3 trimethylation of lysine 9 (H3K9me3) and H4 trimethylation of lysine 20 (H4K20me3) (Schotta et al. 2004).
Histone H3 dimethylation of lysine 9 (H3K9me2)
Uniform H3K9me2 staining was observed as evenly distributed spots all over the autosomal pairs and sex chromosomes in both female and male-derived tammar cells (Fig. 2A). This weak staining did not show any difference between the two X chromosomes in female-derived cells could be observed (Fig. 2Ad). The Y chromosome showed a complete depletion of this modification (Fig. 2Ah).
Status of repressive marks on the inactive X chromosome in tammar wallaby. A Histone H3 dimethylated on lysine 9 (H3K9me2). B Histone H3 trimethylated on lysine 27 (H3K27me3). C Histone H3 trimethylated on lysine 9 (H3K9me3). D Histone H4 trimethylated on lysine 20 (H4K20me3). In each case, the top row shows metaphase spread from female cells and the bottom row shows metaphase spread from male cells. a and e DAPI staining of chromosomes; b and f antibody staining; c and g merged picture (DAPI in red, immunofluorescence in green); d enlarged view of X chromosomes in females (Xa=active X, Xi=inactive X) with line scans for DAPI and histone modification intensities; h enlarged view of X (Xa=active X) and Y chromosomes in males with line scans for DAPI and histone modification intensities. Scale bar represents 10 μm
Histone H3 trimethylation of lysine 27 (H3K27me3)
H3K27me3 staining showed consistent patterns on the arms of autosomal pairs, but not on centromeres (Fig. 2B). On both X chromosomes in female-derived cells, signal was confined to Xq, particularly in the distal region, but one of the X chromosomes appeared slightly brighter (Fig. 2Bd) (>90% of metaphases). This difference could merely reflect the different degrees of chromosome condensation reported previously for the marsupial active and inactive X chromosomes (Graves 1967), and apparent from comparisons of the DAPI staining of the two female X chromosomes (see Alsop et al. 2005). Line scans of intensities of both DAPI and H3K27me3 signals showed that indeed the X chromosome appearing to be slightly enriched in H3K27me3 showed also a slight increase in DAPI intensity (Fig. 2Bd). This suggested that the enrichment in H3K27me3 on the inactive X may be due to its chromatin condensation. In male-derived nuclei, hybridization was also observed on the Y chromosome (Fig. 2Bh).
H3 trimethylation of lysine 9 (H3K9me3)
H3K9me3 produced mainly centromeric staining on all autosomes with some very bright specific signals seen on arms of autosomal pairs 2q, 4p and 6p, 6q (Fig. 2C). The signals on the two X chromosomes in female-derived cells were not consistently different; both showed intense staining on Xp and a band on Xq immediately below the centromere (Fig. 2Cd). Strong hybridization was also observed all over the Y chromosome in male-derived cells (Fig. 2Ch).
H4 trimethylation of lysine 20 (H4K20me3)
The antibody against H4K20me3 revealed more intense binding, but in a very similar pattern to that observed with H3K9me3, especially on the Y chromosome centromeres (Fig. 2D). There were no obvious differences between the two X chromosomes in female-derived cells (Fig. 2Dd).
Discussion
Active marks are lost from the marsupial inactive X chromosome
The eutherian inactive X chromosome is known to lose histone marks associated with transcriptional activation (reviewed by Heard 2005). We now show that the marsupial X, too, is depleted of these marks.
In humans and mice, histone H3 dimethylated at lysine 4 (H3K4me2) has been shown to be distributed throughout most of the interphase nucleus as well as on metaphase chromosomes, but to be absent from the inactive X chromosome (Heard et al. 2001; Boggs et al. 2002; Chaumeil et al. 2002). However, three H3K4me2 hotspots can still be observed on the inactive human X (Boggs et al. 2002). These correspond to the pseudo-autosomal region (PAR) which is not inactivated, a region of the short arm that escapes inactivation (Carrel et al. 1999), and a region near Xq25–26. In mice, too, H3K4me2 is present at the PAR on the distal tip of the inactive X (Chaumeil et al. 2002).
We observed that in female-derived tammar cells, one X had the same H3K4me2 pattern as the single X in males, identifying it as the active X. The other (inactive) X was completely unlabelled, implying that the active mark is lost from the inactive X in marsupials as well as placental mammals. We observed no hotspot of H3K4me2 on the marsupial inactive X, consistent with the absence of a pseudo-autosomal region from the marsupial XY pair (Page et al. 2005). Although preliminary data show that there are regions of the X that escape XCI in female-derived tammar fibroblasts (E. Koina, unpublished), these escapees are evidently not visible as cytogenetic domains. A more detailed gene-by-gene analysis, using chromatin immunoprecipitation assays (ChIP) targeted at specific genes on the tammar X that escape XCI, is being undertaken to clarify whether this histone mark is associated with marsupial escapees.
Underacetylation of histones H3 and H4 is also a feature of the inactive X in both human and mouse female-derived cells (Jeppesen and Turner 1993; Keohane et al. 1996; Heard et al. 2001; Boggs at al. 2002; Chaumeil et al. 2002). However, three discrete regions on the inactive X appear to be normally acetylated, including the PAR, a region located on the short arm (close to Xp22) and a region about half-way down the long arm (at about Xq22), all known to contain genes that escape inactivation (Jeppesen and Turner 1993).
In tammar wallaby metaphase spreads, too, we observed a complete absence of H4 acetylation on the inactive X. Thus the inactive X is underacetylated in marsupials as well as placental mammals. Our results differ somewhat from those of Wakefield et al. (1997), who reported a strong acetylation signal on the NOR-containing short arm of both X chromosomes. The absence of an Xp signal in our experiments may reflect improvement in quality and quantity of metaphases from cytospin preparations and better hybridization techniques that reduce background. Since most of the short arm is heterochromatic, H4 underacetylation is not surprising, although limited binding might be expected around the locus of the repeated ribosomal cistrons, which are active on both X chromosomes.
We also observed underacetylation of histone H3 at lysine 9 (H3K9ac) on the tammar inactive X, except for intense signals in two regions (one proximal, one distal) on the long arm of the inactive X. The hot spot below the centromere contains orthologues of genes in human Xq28, which escapes XCI in humans (Carrel and Willard 2005). In tammar, this region includes G6PD (Koina and Graves 2005) and MeCP2 that at least partially escape XCI in tammar fibroblasts (Koina, preliminary data). The distal region of tammar Xq contains genes with Y homologues in marsupials (Waters et al 2001): ATRX (Y homologue ATRY, Park et al. 2005) and UBE1X (Y homologue UBEIY, Mitchell et al. 1992). These genes also escape XCI in tammar (E. Koina, preliminary data), as do other human X-borne genes with Y homologues (Carrel and Willard 2005).
Thus both histone H3 and H4 appear to be underacetylated on the marsupial inactive X (as they are on the mouse and human inactive X), and H3K9ac (but not H4Kac) binds only to regions in which genes escape XCI.
Status of inactive marks on the marsupial inactive X chromosome
In human and mouse, the inactive X chromosome is known to be enriched in H3K9me2 and H3K27me3, histone modifications associated with transcriptional silencing (Heard et al. 2001; Mermoud et al. 2002; Plath et al. 2003; Silva et al. 2003; Okamoto et al. 2004; Rougeulle et al. 2004). Of the two repressive marks associated with the pericentric constitutive (H3K9me3 and H4K20me3), only H3K9me3 has also been reported to be enriched on a proportion of human inactive X chromosomes (Chadwick and Willard 2004).
In the marsupial tammar wallaby, we could detect no significant enrichment of any of these repressive marks on one of the two X chromosomes on metaphase spreads, nor any significant difference between the two X chromosomes in female-derived cells and the active X in male-derived metaphases. Interestingly, these repressive marks behaved in an opposite way between the short and the long arm of the X chromosome. Indeed, the short arm showed the same pattern of histone modifications as the Y chromosome, a lack of active marks but enrichment in H3K9me3 and H4K20me3. This is consistent with the fact that the short arm of the X chromosome and the Y chromosome carry shared sequences that are mostly heterochromatic (Toder et al. 1997), although the NOR, which is active on both X chromosomes might has been expected to be depleted in repressive marks. The focus of our study here was the long arm of the X chromosome as it represents the common region between human and tammar. In contrast to the short arm, the long arm of the X chromosome showed a global association with H3K9me2 and H3K27me3. But no specific enrichment could be detected for one of the two X chromosomes in female derived-cells.
This absence of enrichment in repressive marks is consistent with the absence of the XIST gene from the marsupial genome (Duret et al. 2006; Hore et al. 2007). Indeed, recruitment of H3K27me3, and probably also of H3K9me2, depends on XIST RNA localization in eutherians (Kohlmaier et al. 2004). In the absence of XIST transcript, these modifications could not be recruited to the inactive X chromosome to help stabilize the inactive state (Silva et al. 2003; Kohlmaier et al. 2004). The lack of enrichment of this inactive marks could render XCI less stable in marsupials than placental mammals, as suggested by the observations of Kaslow and Migeon (1987). This potentially exciting result should be interpreted with some caution, however, since our failure to detect repressive marks on the marsupial inactive X may be a technical issue. Use of antibodies from different suppliers, use of different protocols and reagents, and studies performed on different species can yield inconsistent results in other systems. For instance, whereas Boggs et al. (2002) showed clear enrichment of H3K9me2 on the inactive X in human-derived metaphases, no enrichment on the inactive X in mouse-derived metaphases was observed using either the same antibody (Chaumeil et al. 2002) or a different one (Kohlmaier et al. 2004).
The significance of our observations that repressive marks are absent from the marsupial inactive X must be tested. Their absence on the Xi during mitosis does not necessarily mean that they are not present in interphase. Nevertheless, although they may be present in interphase, their absence through mitosis would imply that they do not behave as real epigenetic marks, as they are thought to act in eutherians. We are currently looking at their status in interphase nuclei, and preliminary results suggest that indeed, unlike in eutherians, H3K27me3 is not stably enriched on the Xi throughout the cell cycle (J. Chaumeil, preliminary data).
In conclusion, our global analysis of histone modifications on metaphase spreads therefore reveals that the marsupial inactive X loses active histone marks, as does the eutherian inactive X. However, our observations suggest that the marsupial inactive X fails to recruit inactive marks, at least in a stable manner. This could be due to the lack of the XIST gene from the marsupial genome that is proposed to recruit repressive marks that are thought to be involved in stabilizing the silencing. This could explain, at least partially, why X chromosome inactivation is less stable in marsupials than in eutherians. Our focus is now shifting from a global examination to determining which histone modifications are associated with specific genes that are either inactive or escape XCI in a complex pattern along the tammar wallaby X chromosome.
Abbreviations
- ATRX, ATRY:
-
alpha thalassaemia/mental retardation syndrome X or Y-linked
- ChIP:
-
chromatin immunoprecipitation
- DAPI:
-
4′,6-diamidino-2-phenylindole dihydrochloride
- FCS:
-
fetal calf serum
- G6PD:
-
glucose-6-phosphate dehydrogenase
- H2AK119ub:
-
histone H2A ubiquitinylation on lysine 119
- H3K27me3:
-
histone H3 trimethylation on lysine 27
- H3K4me2:
-
histone H3 dimethylation on lysine 4
- H3K9ac:
-
histone H3 acetylation on lysine 9
- H3K9me2:
-
histone H3 dimethylation on lysine 9
- H4K20me1:
-
histone H4 monomethylation on lysine 20
- H4Kac:
-
histone H4 acetylation on lysines
- KCM:
-
cytospin buffer containing 10 mM Tris HCl pH 7.6, 120 mM KCl, 20 mM NaCl, 0.5 mM EDTA, 0.1% Triton; pH 7.5
- Lnx3:
-
ligand of NUMB-protein X 3 (also known as PDZRN3 or SEMCAP3)
- MeCP2:
-
methyl CpG binding Protein 2
- Mya:
-
million years ago
- NOR:
-
nucleolus organizer region
- PAR:
-
pseudo-autosomal region
- UBE1X, UBE1Y:
-
Ubiquitin-activating enzyme E1, X or Y-linked
- XCI:
-
X chromosome inactivation
- XIC:
-
X inactivation centre
- Xist:
-
X-inactive specific transcript
- Xp:
-
short arm of the X chromosome
- Xq:
-
long arm of the X chromosome
References
Alsop AE, Miethke P, Rofe R et al (2005) Characterizing the chromosomes of the Australian model marsupial Macropus eugenii (tammar wallaby). Chromosome Res 3:627–636
Berger SL (2007) The complex language of chromatin regulation during transcription. Nature 447:19337–19344
Boggs BA, Cheung P, Heard E et al (2002) Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat Genet 30:73–76
Brown CJ, Ballabio A, Rupert JL et al (1991) A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349:38–44
Carrel L, Willard HF (2005) X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434:400–404
Carrel L, Cottle AA, Goglin KC, Willard HF (1999) A first-generation X-inactivation profile of the human X chromosome. Proc Natl Acad Sci USA 96:14440–14444
Chadwick BP, Willard HF (2004) Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc Natl Acad Sci USA 101:17450–17455
Chaumeil J, Okamoto I, Guggiari M, Heard E (2002) Integrated kinetics of X chromosome inactivation in differentiating embryonic stem cells. Cytogenet Genome Res 99:75–84
Chaumeil J, Okamoto I, Heard E (2004) X-chromosome inactivation in mouse embryonic stem cells: analysis of histone modifications and transcriptional activity using immunofluorescence and FISH. Methods Enzymol 376:405–419
Chureau C, Prissette M, Bourdet A et al (2002) Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine. Genome Res 12:894–908
Cooper DW (1971) Directed genetic change model for X chromosome inactivation in eutherian mammals. Nature 230:292–294
Cooper DW, Johnston PG, Graves JAM (1993) X-inactivation in marsupials and monotremes. Semin Dev Biol 4:117–128
Costanzi C, Pehrson JR (1998) Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393:599–601
de Napoles M, Mermoud JE, Wakao R et al (2004) Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7:663–676
Deakin JE, Hore TA, Koina E, Graves JAM (2008) Dosage Compensation in the multiple X chromosomes of the platypus. BMC Genomics. PLoS Genet 4(7):e1000140, July 25.
Duret L, Chureau C, Samain S, Weissenbach J, Avner P (2006) The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 312:1653–1655
Fang J, Chen T, Chadwick B, Li E, Zhang Y (2004) Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J Biol Chem 279:52812–52815
Graves JAM (1967) DNA synthesis in chromosomes of cultured leucocytes from two marsupial species. Exp Cell Res 46:37–57
Graves JAM, Gartler SM (1986) Mammalian X chromosome inactivation: testing the hypothesis of transcriptional control. Somat Cell Mol Genet 12:275–280
Graves JAM, Dawson GW (1988) The relationship between position and expression of genes on the kangaroo X chromosome suggests a tissue-specific spread of inactivation from a single control site. Genet Res 51:103–109
Heard E (2005) Delving into the diversity of facultative heterochromatin: The epigenetics of the inactive X chromosome. Curr Opin Genet Dev 15:482–489
Heard E, Disteche CM (2006) Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev 20:1848–1867
Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL (2001) Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell 107:727–738
Hernandez-Munoz I, Lund AH, van der Stoop P et al (2005) Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci USA 102:7635–7640
Hore TA, Koina E, Wakefield MJ, Graves JAM (2007) The region homologous to the X-chromosome inactivation centre has been disrupted in marsupial and monotreme mammals. Chromosome Res 15:147–161
Huynh KD, Lee JT (2003) Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature 426:857–862
Itoh Y, Melamed E, Yang X et al (2007) Dosage compensation is less effective in birds than in mammals. J Biol 6:2
Jeppesen P, Turner BM (1993) The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell 74:281–289
Kaslow DC, Migeon BR (1987) DNA methylation stabilizes X chromosome inactivation in eutherians but not in marsupials: evidence for multistep maintenance of mammalian X dosage compensation. Proc Natl Acad Sci USA 84:6210–6214
Keohane AM, O’Neill LP, Belyaev ND, Lavender JS, Turner BM (1996) X-Inactivation and histone H4 acetylation in embryonic stem cells. Dev Biol 180:618–630
Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A (2004) A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol 2:E171
Koina E, Wakefield MJ, Walcher C et al (2005) Isolation, X location and activity of the marsupial homologue of SLC16A2, an XIST-flanking gene in eutherian mammals. Chromosome Res 13:687–698
Kouzarides T (2007) Chromatin modifications and their function. Cell 128:693–705
Lock LF, Takagi N, Martin GR (1987) Methylation of the Hprt gene on the inactive X occurs after chromosome inactivation. Cell 48:39–46
Loebel DA, Johnston PG (1996) Methylation analysis of a marsupial X-linked CpG island by bisulfite genomic sequencing. Genome Res 6:114–123
Lyon MF (1961) Gene action in the X chromosome of the mouse (Mus musculus L.). Nature 190:372–373
Mermoud JE, Costanzi C, Pehrson JR, Brockdorff N (1999) Histone macroH2A1.2 relocates to the inactive X chromosome after initiation and propagation of X-inactivation. J Cell Biol 147:1399–1408
Mermoud JE, Popova B, Peters AH, Jenuwein T, Brockdorff N (2002) Histone H3 lysine 9 methylation occurs rapidly at the onset of random X chromosome inactivation. Curr Biol 12:247–251
Mitchell MJ, Woods DR, Wilcox SA, Graves JAM, Bishop CE (1992) Marsupial Y chromosome encodes a homologue of the mouse Y-linked candidate spermatogenesis gene Ubely. Nature 359:528–531
Nightingale KP, O’Neill LP, Turner BM (2006) Histone modifications: signalling receptors and potential elements of a heritable epigenetic code. Curr Opin Genet Dev 16:125–136
Norris DP, Brockdorff N, Rastan S (1991) Methylation status of CpG-rich islands on active and inactive mouse X chomosomes. Mamm Genome 1:78–83
Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E (2004) Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303:644–649
Okamoto I, Arnaud D, Le Baccon P et al (2005) Evidence for de novo imprinted X-chromosome inactivation independent of meiotic inactivation in mice. Nature 438:369–373
Page J, Berríos S, Parra MT et al (2005) The program of sex chromosome pairing in meiosis is highly conserved across marsupial species: implications for sex chromosome evolution. Genetics 170:793–799
Park DJ, Pask AJ, Huynh K, Harley VR, Renfree MB, Graves JAM (2005) Characterisation of the marsupial-specific ATRY gene: implications for the evolution of male-specific function. Gene 362:29–36
Piper AA, Bennett AM, Noyce L, Swanton MK, Cooper DW (1993) Isolation of a clone partially encoding hill kangaroo X-linked hypoxanthine phosphoribosyltransferase: sex differences in methylation in the body of the gene. Somat Cell Mol Genet 19:141–159
Plath K, Fang J, Mlynarczyk-Evans SK et al (2003) Role of histone H3 lysine 27 methylation in X inactivation. Science 300:131–135
Rastan S, Robertson EJ (1985) X-chromosome deletions in embryo-derived (EK) cell lines associated with lack of X-chromosome inactivation. J Embryol Exp Morphol 90:379–388
Rougeulle C, Chaumeil J, Sarma K et al (2004) Differential histone H3 Lys-9 and Lys-27 methylation profiles on the X chromosome. Mol Cell Biol 24:5475–5484
Sado T, Okano M, Li E, Sasaki H (2004) De novo DNA methylation is dispensable for the initiation and propagation of X chromosome inactivation. Development 131:975–982
Schotta G, Lachner M, Sarma K et al (2004) A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 18:1251–1262
Sharman GB (1971) Late DNA replication in the paternally derived X chromosome of female kangaroos. Nature 230:231–232
Shevchenko AI, Zakharova IS, Elisaphenko EA et al (2007) Genes flanking Xist in mouse and human are separated on the X chromosome in American marsupials. Chromosome Res 15:127–136
Silva J, Mak W, Zvetkova I et al (2003) Establishment of histone H3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell 4:481–495
Takagi N, Sasaki M (1975) Preferential inactivation of the paternally derived X-chromosome in the extraembryonic membranes of the mouse. Nature 256:640–642
Toder R, Wienberg J, Voullaire L, O’Brien PC, Maccarone P, Graves JAM (1997) Shared DNA sequences between the X and Y chromosomes in the tammar wallaby—evidence for independent additions to eutherian and marsupial sex chromosomes. Chromosoma 106:94–98
Turner BM (2007) Defining an epigenetic code. Nat Cell Biol 9:2–6
VandeBerg JL, Robinson ES, Samollow PB, Johnston PG (1987) X-linked gene expression and X-chromosome inactivation: marsupials, mouse, and man compared. Isozymes Curr Top Biol Med Res 15:225–253
Veyrunes F, Waters PD, Miethke P et al (2008) Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res 18:965–973
Wakefield MJ, Keohane AM, Turner BM, Graves JAM (1997) Histone underacetylation is an ancient component of mammalian X chromosome inactivation. Proc Natl Acad Sci USA 94:9665–9668
Waters PD, Duffy B, Frost CJ, Delbridge ML, Graves JAM (2001) The human Y chromosome derives largely from a single autosomal region added to the sex chromosomes 80–130 million years ago. Cytogenet Cell Genet 92:74–79
Acknowledgements
We acknowledge funding to J.A.M.G. from The Australian Research Council. J.C. was funded by the French ‘Association pour la Recherche sur le Cancer’.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Herbert Macgregor.
Edda Koina and Julie Chaumeil contributed equally to this work.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Koina, E., Chaumeil, J., Greaves, I.K. et al. Specific patterns of histone marks accompany X chromosome inactivation in a marsupial. Chromosome Res 17, 115–126 (2009). https://doi.org/10.1007/s10577-009-9020-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-009-9020-7