Abstract
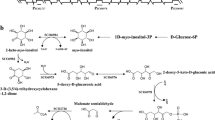
The GATA-type transcriptional repressor structural gene SRE1 was isolated from both the genomic DNA and mRNA of the marine yeast Aureobasidium pullulans HN6.2 by inverse PCR and RACE. An open reading frame (ORF) of 1,002 bp encoding a 334 amino acid protein (a calculated isoelectric point: 8.6) with a calculated molecular weight of 35.1 kDa was characterized. The corresponding gene had one single intron of 51 bp, and in its promoter two putative 5′-HGATAR-3′ sequences could be recognized. The deduced protein from the cloned gene contained two conserved zinc-finger domains [Cys-(X2)-Cys-(X17)-Cys-(X2)-Cys)], nine sequences of Ser(Thr)-Pro-X-X which was characteristics of the regulator, and one cysteine-rich central domain which was located between the two zinc fingers. The SRE1 gene in A. pullulans HN6.2 was disrupted by integrating the hygromycin B phosphotransferase gene into the ORF of the SRE1 gene using homologous recombination. Two hundreds of the disruptants (Δsre1) (one of them was named R6) obtained still synthesized both intracellular and extracellular siderophores in the presence of added Fe3+ and the expression of the SidA gene encoding l-ornithine N5-oxygenase in the disruptant R6 was also partially derepressed in the presence of added Fe3+. The colonies of the disruptant R6 grown on the iron-replete medium with 1.5 and 2.0 mM Fe3+ and also with 1.5 mM Fe2+ became brown. In contrast, A. pullulans HN6.2 could not grow in the iron-replete medium with 1.5 mM and 2.0 mM Fe3+. The brown-colored colonies of the disruptant R6 also had high level of siderophore and iron.






Similar content being viewed by others
References
An Z, Zhao Q, McEvoy J, Yuan WM, Markley JL, Leong SA (1996) The second finger of Urbs1 is required for iron-mediated repression of sid1 in Ustilago maydis. Proc Nat Acad Sci USA 94:5882–5887
An Z, Mei B, Yuan WM, Leong SA (1997) The distal GATA sequences of the sid1 promoter of Ustilago maydis mediate iron repression of siderophore production and interact directly with Urbs1, a GATA family transcription factor. EMBO J 16:1742–1750
Baakza A, Vala AK, Dave BP, Dube HC (2004) A comparative study of siderophore production by fungi from marine and terrestrial habitats. J Exp Mar Biol Ecol 311:1–9
Bradford MM (1976) A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72:248–253
Chi ZM, Fang Wang F, Chi Z, Yue L, Liu GL, Zhang T (2009) Bioproducts from Aureobasidium pullulans, a biotechnologically important yeast. Appl Microbiol Biotechnol 82:793–804
Chi ZM, Liu GL, Zhao SF, Li J, Peng Y (2010) Marine yeasts as biocontrol agents and producers of bio-products. Appl Microbiol Biotechnol 86:1227–1241
Chi Z, Wang XX, Ma ZC, Buzdar MA, Chi ZM (2012) The unique role of siderophore in marine-derived Aureobasidium pullulans HN6.2. Biomet 25:219–230
Gauthier GM, Sullivan TD, Gallardo SS, Brandhorst TT, Wymelenberg AJV, Cuomo CA, Suen G, Currie CR, Klein BS (2010) SREB, a GATA transcription factor that directs disparate fates in Blastomyces dermatitidis including morphogenesis and siderophore biosynthesis. PLoS Pathog 6:1–16
Haas H (2003) Molecular genetics of fungal siderophore biosynthesis and uptake: the role of siderophores in iron uptake and storage. Appl Microbiol Biotechnol 62:316–330
Haas H, Angermayr K, Stoffler G (1997) Molecular analysis of a Penicillium chrysogenum GATA factor encoding gene (sreP) exhibiting significant homology to the Ustilago maydis urbs1 gene. Gene 184:33–37
Haas H, Zadra I, Stoeffler G, Angermayr K (1999) The Aspergillus nidulans GATA factor SREA is involved in regulation of siderophore biosynthesis and control of iron uptake. J Biol Chem 274:4613–4619
Holinsworth B, Martin JD (2009) Siderophore production by marine-derived fungi. Biomet 22:625–632
Johnson L (2008) Iron and siderophores in fungal-host interactions. Mycol Res 112:170–183
Jung WH, Sham A, White R, Kronstad JW (2006) Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol 4:2282–2295
Labbe S, Pelletier B, Mercier A (2007) Iron homeostasis in the fission yeast Schizosaccharomyces pombe. Biomet 20:523–537
Leong SA, Winkelmann G (1998) Molecular biology of iron transport in fungi. Met Ions Biol Syst 35:147–186
Liu GL, Wang DS, Wang LF, Zhao SF, Chi ZM (2011) Mig1 is involved in mycelial formation and expression of the genes encoding extracellular enzymes in Saccharomycopsis fibuligera A11. Fung Genet Biol 48:904–913
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCt method. Meth 25:402–408
Mei B, Budde AD, Leong SA (1993) sid1, a gene initiating siderophore biosynthesis in Ustilago maydis: molecular characterization, regulation by iron, and role in phytopathogenicity. Proc Nat Acad Sci USA 90:903–907
Ni X, Yue L, Chi ZM, Li J, Wang X, Madzak C (2009) Alkaline protease gene cloning from the marine yeast Aureobasidium pullulans HN2-3 and the protease surface display on Yarrowia lipolytica for bioactive peptide production. Mar Biotechnol 11:81–89
Oberegger H, Schoeser M, Zadra I, Abt B, Haas H (2001) SREA is involved in regulation of siderophore biosynthesis, utilization and uptake in Aspergillus nidulans. Mol Microbiol 41:1077–1089
Pelletier B, Trott A, Morano KA, Labbe S (2005) Functional characterization of the iron-regulatory transcription factor Fep1 from Schizosaccharomyces pombe. J Biol Chem 280:25146–25161
Philpott CC (2006) Iron uptake in fungi: a system for every source. Biochim Biophys Acta 1763:636–645
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Beijing, pp 367–370 (Chinese translated ed)
Schrettl M, Kim HS, Eisendle M, Kragl C, Nierman WC, Heinekamp T, Werner ER, Jacobsen I, Illmer P, Yi H, Brakhage AA, Haas H (2008) SreA-mediated iron regulation in Aspergillus fumigatus. Mole Microbiol 70:27–43
Schrettl M, Beckmann N, Varga J, Heinekamp T, Jacobsen ID, Jochl C, Moussa TA, Wang S, Gsaller F, Blatzer M, Werner ER, Niermann WC, Brakhage AA, Haas H (2010) HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog 6:e1001124
Tamarit J, Irazusta V, Moreno-Cermeno A, Ros J (2006) Colorimetric assay for the quantitation of iron in yeast. Anal Biochem 351:149–151
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tomlinson G, Crujckshank WH, Viswanatha T (1971) Sensitivity of substituted hydroxylamines to determination by iodine oxidation. Anal Biochem 44:70–679
Wang W, Chi ZM, Chi Z, Li J, Wang XH (2009a) Siderophore production by the marine-derived Aureobasidium pullulans and its antimicrobial activity. Bioresour Technol 100:2639–2641
Wang W, Chi Z, Liu G, Buzdar MA, Chi ZM, Gu Q (2009b) Chemical and biological characterization of siderophore produced by the marine-derived Aureobasidium pullulans HN6.2 and its antibacterial activity. Biomet 22:965–972
Watanabe H, Hatakeyama N, Sakurai H, Uchimiya H, Sato T (2008) Isolation of industrial strains of Aspergillus oryzae lacking ferrichrysin by disruption of the dffA gene. J Biosci Bioeng 106:488–492
Wu JCF, Hamada M (2000) Experiments planning, analysis and parameter design optimization. Wiley, New York, pp 23–34
Yuan Y, Guo X, Xiuping He, Zhang B, Liu S (2004) Construction of a high-biomass, iron-enriched yeast strain and study ondistribution of iron in the cells of Saccharomyces cerevisiae. Biotechnol Lett 26:311–315
Zhou LW, Haas H, Marzluf GA (1998) Isolation and characterization of a new gene, sre, which encodes a GATA-type regulatory protein that controls iron transport in Neurospora crassa. Mol Gen Genet 259:532–540
Acknowledgments
This work was supported by Grant 31070029 from National Natural Science Foundation of China.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chi, Z., Wang, XX., Geng, Q. et al. Role of a GATA-type transcriptional repressor Sre1 in regulation of siderophore biosynthesis in the marine-derived Aureobasidium pullulans HN6.2. Biometals 26, 955–967 (2013). https://doi.org/10.1007/s10534-013-9672-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10534-013-9672-9