Abstract
Objectives
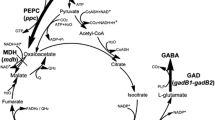
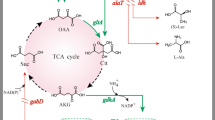
To enhance γ-aminobutyric acid (GABA) production in recombinant Corynebacterium glutamicum, metabolic engineering strategies were used to improve the supply of the GABA precursor, l-glutamate.
Results
C. glutamicum ATCC13032 co-expressing two glutamate decarboxylase genes (gadB1 and gadB2) was constructed in a previous study Shi et al. (J Ind Microbiol Biotechnol 40:1285–1296, 2013) to synthesize GABA from endogenous l-glutamate. To improve its l-glutamate supply, new strains were constructed here. First, the odhA and pyc genes were deleted separately. Then, a gadB1–gadB2 co-expression plasmid was transferred into ΔodhA, Δpyc, and ATCC13032, resulting in recombinant strains SNW201, SNW202, and SNW200, respectively. After fermenting for 72 h, GABA production increased to 29.5 ± 1.1 and 24.9 ± 0.7 g/l in SNW201 and SNW202, respectively, which was significantly higher than that in SNW200 (19.4 ± 2.6 g/l). The GABA conversion ratios of SNW201 and SNW202 reached 0.98 and 0.96 mol/mol, respectively.
Conclusion
The recombinant strains SNW201 and SNW202 can be used as candidates for GABA production.





Similar content being viewed by others
References
Asakura Y, Kimura E, Usuda Y, Kawahara Y, Matsui K, Osumi T, Nakamatsu T (2007) Altered metabolic flux due to deletion of odhA causes l-glutamate overproduction in Corynebacterium glutamicum. Appl Environ Microbiol 73:1308–1319
Hu JY, Tan YZ, Li YY, Hu XQ, Xu DQ, Wang XY (2013) Construction and application of an efficient multiple-gene-deletion system in Corynebacterium glutamicum. Plasmid 70:303–313
Hu JY, Li YY, Zhang HL, Tan YZ, Wang XY (2014) Construction of a novel expression system for use in Corynebacterium glutamicum. Plasmid 75:18–26
Li HX, Qiu T, Gao DD, Cao YS (2010a) Medium optimization for production of gamma-aminobutyric acid by Lactobacillus brevis NCL912. Amino Acids 38:1439–1445
Li HX, Qiu T, Huang GD, Cao YS (2010b) Production of gamma-aminobutyric acid by Lactobacillus brevis NCL912 using fed-batch fermentation. Microb Cell Fact 9:85
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–∆∆CT method. Methods 25:402–408
Park KB, Oh SH (2007) Cloning, sequencing and expression of a novel glutamate decarboxylase gene from a newly isolated lactic acid bacterium, Lactobacillus brevis OPK-3. Bioresour Technol 98:312–319
Shi F, Li YX (2011) Synthesis of γ-aminobutyric acid by expressing Lactobacillus brevis-derived glutamate decarboxylase in the Corynebacterium glutamicum strain ATCC 13032. Biotechnol Lett 33:2469–2474
Shi F, Jiang JJ, Li YF, Li YX, Xie YL (2013) Enhancement of γ-aminobutyric acid production in recombinant Corynebacterium glutamicum by co-expressing two glutamate decarboxylase genes from Lactobacillus brevis. J Ind Microbiol Biotechnol 40:1285–1296
Shi F, Xie YL, Jiang JJ, Wang NN, Li YF, Wang XY (2014) Directed evolution and mutagenesis of glutamate decarboxylase from Lactobacillus brevis Lb85 to broaden the range of its activity toward a near-neutral pH. Enzyme Microb Technol 61–62:35–43
Shiio I, Ozaki H (1970) Regulation of nicotinamide adenine dinucleotide phosphate-specific glutamate dehydrogenase from Brevibacterium flavum, a glutamate-producing bacterium. J Biochem 68:633–647
Shirai T, Fujimura K, Furusawa C, Nagahisa K, Shioya S, Shimizu H (2007) Study on roles of anaplerotic pathways in glutamate overproduction of Corynebacterium glutamicum by metabolic flux analysis. Microb Cell Fact 6:19
Yao WJ, Deng XZ, Zhong H, Liu M, Zheng P, Sun ZH, Zhang Y (2009) Double deletion of dtsR1 and pyc induce efficient l-glutamate overproduction in Corynebacterium glutamicum. J Ind Microbiol Biotechnol 36:911–921
Acknowledgments
The authors thank the ‘‘Program of State Key Laboratory of Food Science and Technology” (contract no. SKLF-ZZB-201405) and ‘‘Fundamental Research Funds for the Central Universities” (contract no. JUSRP51303A) for financial support.
Supporting information
Supplementary Table 1—Primers used in this study. The restriction sites are in boldface. The start and stop codons are underlined.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, N., Ni, Y. & Shi, F. Deletion of odhA or pyc improves production of γ-aminobutyric acid and its precursor l-glutamate in recombinant Corynebacterium glutamicum . Biotechnol Lett 37, 1473–1481 (2015). https://doi.org/10.1007/s10529-015-1822-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-015-1822-4