Abstract
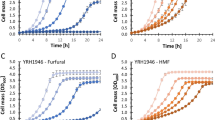
Genome shuffling was used to obtain Pachysolen tannophilus mutants with improved tolerance to inhibitors in hardwood spent sulfite liquor (HW SSL). Genome shuffled strains (GHW301, GHW302 and GHW303) grew at higher concentrations of HW SSL (80 % v/v) compared to the HW SSL UV mutant (70 % v/v) and the wild-type (WT) strain (50 % v/v). In defined media containing acetic acid (0.70–0.90 % w/v), GHW301, GHW302 and GHW303 exhibited a shorter lag compared to the acetic acid UV mutant, while the WT did not grow. Genome shuffled strains produced more ethanol than the WT at higher concentrations of HW SSL and an aspen hydrolysate. To identify the genetic basis of inhibitor tolerance, whole genome sequencing was carried out on GHW301, GHW302 and GHW303 and compared to the WT strain. Sixty single nucleotide variations were identified that were common to all three genome shuffled strains. Of these, 40 were in gene sequences and 20 were within 5 bp–1 kb either up or downstream of protein encoding genes. Based on the mutated gene products, mutations were grouped into functional categories and affected a variety of cellular functions, demonstrating the complexity of inhibitor tolerance in yeast. Sequence analysis of UV mutants (UAA302 and UHW303) from which GHW301, GHW302 and GHW303 were derived, confirmed the success of our cross-mating based genome shuffling strategy. Whole-genome sequencing analysis allowed identification of potential gene targets for tolerance to inhibitors in lignocellulosic hydrolysates.






Similar content being viewed by others
References
Aasland R, Gibson TJ, Stewart AF (1995) The PHD finger: implications for chromatin mediated transcriptional regulation. Trends Biochem Sci 20:56–59
Allen SA, Clark W, McCaffery JM, Cai Z, Lanctot A, Slininger PJ, Liu ZL, Gorsich SW (2010) Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnol Biofuels 3:2–11
Almeida B, Ohlmeier S, Almeida AJ, Madeo F, Leao C, Rodrigues F, Ludovico P (2009) Yeast protein expression profile during acetic acid-induced apoptosis indicates causal involvement of the TOR pathway. Proteomics 9:720–732
Andre B (1995) An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast 11:1575–1611
Ask M, Bettiga M, Duraiswamy VR, Olsson L (2013) Pulsed addition of HMF and furfural to batch-grown xylose-utilizing Saccharomyces cerevisiae results in different physiological responses in glucose and xylose consumption phase. Biotechnol Biofuels 6:181–195
Ask M, Bettiga M, Mapelli V, Olsson L (2013) The influence of HMF and furfural on redox-balance and energy-state of xylose-utilizing Saccharomyces cerevisiae. Biotechnol Biofuels 6:22–27
Bajwa PK, Shireen T, D’Aoust F, Pinel D, Martin VJJ, Trevors JT, Lee H (2009) Mutants of the pentose-fermenting yeast Pichia stipitis with improved tolerance to inhibitors in hardwood spent sulfite liquor. Biotechnol Bioeng 104:892–900
Bajwa PK, Pinel D, Martin VJJ, Trevors JT, Lee H (2010) Strain improvement of the pentose-fermenting yeast Pichia stipitis by genome shuffling. J Microbiol Methods 81:179–186
Bajwa PK, Phaenark C, Grant N, Zhang X, Paice M, Martin VJJ, Trevors JT, Lee H (2011) Ethanol production from selected lignocellulosic hydrolysates by genome shuffled strains of Scheffersomyces stipitis. Bioresour Technol 102:9965–9969
Bajwa PK, Harner NK, Richardson TL, Sindu S, Habash MB, Trevors JT, Lee H (2013) Genome shuffling protocol for the pentose-fermenting yeast Scheffersomyces stipitis. In: Gupta VK, Tuohy MG, Ayyachamy M, Turner KM, O’Donovan A (eds) Laboratory protocols in fungal biology: current methods in fungal biology. Springer, New York, pp 447–454
Bajwa PK, Ho CY, Chan CK, Martin VJJ, Trevors JT, Lee H (2013) Transcriptional profiling of Saccharomyces cerevisiae T2 cells upon exposure to hardwood spent sulphite liquor: comparison to acetic acid, furfural and hydroxymethylfurfural. Antonie Van Leeuwenhoek 103:1281–1295
Barbosa MDS, Lee H, Collins-Thompson DL (1990) Additive effects of alcohols, their acidic by-products, and temperature on the yeast Pachysolen tannophilus. Appl Environ Microbiol 56:545–550
Barbosa MDS, Lee H, Schneider H, Forsberg CW (1990) Temperature mediated changes of D-xylose metabolism in the yeast Pachysolen tannophilus. FEMS Microbiol Lett 72:35–40
Bauer BE, Rossington D, Mollapour M, Mamnun Y, Kuchler K, Piper PW (2003) Weak organic acid stress inhibits aromatic amino acid uptake by yeast, causing a strong influence of amino acid auxotrophies on the phenotypes of membrane transporter mutants. Eur J Biochem 270:3189–3195
Bicho PA, Runnals PL, Cunningham JD, Lee H (1988) Induction of xylose reductase and xylitol dehydrogenase activities in Pachysolen tannophilus and Pichia stipitis on mixed sugars. Appl Environ Microbiol 54:50–54
Biot-Pelletier D, Martin VJJ (2014) Evolutionary engineering by genome shuffling. Appl Microbiol Biotechnol 98:3877–3887
Bose S, Dutko JA, Zitomer RS (2005) Genetic factors that regulate the attenuation of the general stress response of yeast. Genetics 169:1215–1226
Choi Y, Sims GE, Murphy S, Miller JR, Chan AP (2012) Predicting the functional effect of amino acid substitutions and indels. PLoS One 7:1–13
Clark T, Wedlock N, James AP, Deverell K, Thornton RJ (1986) Strain improvement of xylose-fermenting yeast Pachysolen tannophilus by hybridization of two mutant strains. Biotechnol Lett 8:801–806
Davierwala AP, Haynes J, Li ZJ, Brost RL, Robinson MD, Yu L, Mnaimneh S, Ding HM, Zhu HW, Chen YQ, Cheng X, Brown GW, Boone C, Andrews BJ, Hughes TR (2005) The synthetic genetic interaction spectrum of essential genes. Nat Genet 37:1147–1152
Delley PA, Hall MN (1999) Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J Cell Biol 147:163–174
Dinh TN, Nagahisa K, Yoshikawa K, Hirasawa T, Furusawa C, Shimizu H (2009) Analysis of adaptation to high ethanol concentration in Saccharomyces cerevisiae using DNA microarray. Bioprocess Biosyst Eng 32:681–688
Dunlop MJ (2011) Engineering microbes for tolerance to next-generation biofuels. Biotechnol Biofuels 4:32–40
Furukawa K, Kitano H, Mizoguchi H, Hara S (2004) Effect of cellular inositol content on ethanol tolerance of Saccharomyces cerevisiae in sake brewing. J Biosci Bioeng 98:107–113
Gaber RF, Kielland-Brandt MC, Fink GR (1990) HOL1 mutations confer novel ion transport in Saccharomyces cerevisiae. Mol Cell Biol 10:643–652
Gandhi M, Goode BL (2008) Coronin: the double-edged sword of actin dynamics. Subcell Biochem 48:72–87
Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11:4241–4257
Giudici P, Solieri L, Pulvirenti AM, Cassanelli S (2005) Strategies and perspectives for genetic improvement of wine yeasts. Appl Microbiol Biotechnol 66:622–628
Gorsich SW, Dien BS, Nichols NN, Slininger PJ, Liu ZL, Skory CD (2006) Tolerance to furfural-induced stress is associated with pentose phosphate pathway genes ZWF1, GND1, RPE1, and TKL1 in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 71:339–349
Gorsich SW, Slininger PJ, McCaffery JM (2006b) The fermentation inhibitor furfural causes cellular damage to Saccharomyces cerevisiae. In: Biotechnology for fuels and chemical symposium proceedings, Paper no. 4–17
Hao XC, Yang XS, Wan P, Tian S (2013) Comparative proteomic analysis of a new adaptive Pichia stipitis strain to furfural, a lignocellulosic inhibitory compound. Biotechnol Biofuels 6:34–42
Harner NK, Bajwa PK, Habash MB, Trevors JT, Austin GD, Lee H (2014) Mutants of the pentose-fermenting yeast Pachysolen tannophilus tolerant to hardwood spent sulfite liquor and acetic acid. Antonie Van Leeuwenhoek 105:29–43
Harner NK, Wen X, Bajwa PK, Austin GD, Ho CY, Habash MB, Trevors JT, Lee H (2015) Genetic improvement of native xylose-fermenting yeasts for ethanol production. J Ind Microbiol Biotechnol 42:1–20
Hashikawa N, Mizukami Y, Imazu H, Sakurai H (2006) Mutated yeast heat shock transcription factor activates transcription independently of hyperphosphorylation. J Biol Chem 281:3936–3942
James AP, Zahab DM (1982) A genetic system for Pachysolen tannophilus, a pentose-fermenting yeast. J Gen Microbiol 128:2297–2301
James AP, Zahab DM (1983) The construction and genetic analysis of polyploids and aneuploids of the pentose-fermenting yeast, Pachysolen tannophilus. J Gen Microbiol 129:2489–2494
Jeffries TW (1984) Mutants of Pachysolen tannophilus showing enhanced rates of growth and ethanol formation from d-xylose. Enzyme Microb Technol 6:254–258
Jiang H, Xie YQ, Houston P, Stemkehale K, Mortensen UH, Rothstein R, Kodadek T (1996) Direct association between the yeast Rad51 and Rad54 recombination proteins. J Biol Chem 271:33181–33186
Kim DR, Gidvani RD, Ingalls BP, Duncker BP, McConkey BJ (2011) Differential chromatin proteomics of the MMS-induced DNA damage response in yeast. Proteome Sci 9:62–75
Kim HS, Kim NR, Kim W, Choi W (2012) Insertion of transposon in the vicinity of SSK2 confers enhanced tolerance to furfural in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 95:531–540
Krogan NJ, Cagney G, Yu HY, Zhong GQ, Guo XH, Ignatchenko A, Li J, Pu SY, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St Onge P, Ghanny S, Lam MHY, Butland G, Altaf-Ui AM, Kanaya S, Shilatifard A, O’Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440:637–643
Künzler M, Trueheart J, Sette C, Hurt E, Thorner J (2001) Mutations in the YRB1 gene encoding yeast Ran-binding-protein-1 that impair nucleocytoplasmic transport and suppress yeast mating defects. Genetics 157:1089–1105
Kuranda MJ, Robbins PW (1991) Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J Biol Chem 266:19758–19767
Larsson S, Palmqvist E, Hahn-Hagerdal B, Tengborg C, Stenberg K, Zacchi G, Nilvebrant NO (1999) The generation of fermentation inhibitors during dilute acid hydrolysis of softwood. Enzyme Microb Technol 24:151–159
Lee H, James AP, Zahab DM, Mahmourides G, Maleszka R, Schneider H (1986) Mutants of Pachysolen tannophilus with improved production of ethanol D-xylose. Appl Environ Microbiol 51:1252–1258
Li BZ, Yuan YJ (2010) Transcriptome shifts in response to furfural and acetic acid in Saccharomyces cerevisiae. Appl Microbiol Boitechnol 86:1915–1924
Ligthelm ME, Prior BA, du Preez JC, Brandt V (1988) An investigation of D-{1-13C} xylose metabolism in Pichia stipitis under aerobic and anaerobic conditions. Appl Microbiol Biotechnol 28:293–296
Lin FM, Qiao B, Yuan YJ (2009) Comparative proteomic analysis of tolerance and adaptation of ethanologenic Saccharomyces cerevisiae to furfural, a lignocellulosic inhibitory compound. Appl Environ Microbiol 75:3765–3776
Lindberg L, Santos AXS, Riezman H, Olsson L, Bettiga M (2013) Lipidomic profiling of Saccharomyces cerevisiae and Zygosaccharomyces bailii reveals critical changes in lipid composition in response to acetic acid stress. PLoS One 8:1–12
Liu ZL (2011) Molecular mechanisms of yeast tolerance and in situ detoxification of lignocellulose hydrolysates. Appl Microbiol Biotechnol 90:809–825
Liu ZL, Slininger PJ (2006) Transcriptome dynamics of ethanologenic yeast in response to 5-hydroxymethylfurfural stress related to biomass conversion to ethanol. In: Mendez-Vilas A (ed) Modern multidisciplinary applied microbiology: exploiting microbes and their interactions. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, pp 679–684
Liu ZL, Ma MG, Song MZ (2009) Evolutionarily engineered ethanologenic yeast detoxifies lignocellulosic biomass conversion inhibitors by reprogrammed pathways. Mol Genet Genomics 282:233–244
Liu XY, Kaas RS, Jensen PR, Workman M (2012) Draft genome sequence of the yeast Pachysolen tannophilus CBS 4044/NRRL Y-2460. Eukaryot Cell 11:827
Lohmeier-Vogel EM, Sopher CR, Lee H (1998) Intracellular acidification as a mechanism for the inhibition by acid hydrolysis-derived inhibitors of xylose fermentation by yeasts. J Ind Microbiol Biotechnol 20:75–81
Ma MG, Liu LZ (2010) Quantitative transcription dynamic analysis reveals candidate genes and key regulators for ethanol tolerance in Saccharomyces cerevisiae. BMC Microbiol 10:169–188
Ma MG, Liu ZL (2010) Comparative transcriptome profiling analyses during the lag phase uncover YAP1, PDR1, PDR3, RPN4, and HSF1 as key regulatory genes in genomic adaptation to the lignocellulose derived inhibitor HMF for Saccharomyces cerevisiae. BMC Genomics 11:660–678
MacPherson S, Larochelle M, Turcotte B (2006) A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev 70:583–604
Maleszka R, Schneider H (1982) Concurrent production and consumption of ethanol by cultures of Pahcysolen tannophilus growing on d-xylose. Appl Environ Microbiol 44:909–912
Mira NP, Becker JD, Sá-Correia I (2010) Genomic expression program involving the Haa1p-regulon in Saccharomyces cerevisiae response to acetic acid. OMICS 14:587–601
Mira NP, Palma M, Guerreiro JF, Sá-Correia I (2010) Genome-wide identification of Saccharomyces cerevisiae genes required for tolerance to acetic acid. Microb Cell Fact 9:79–91
Moran GR (2005) 4-hydroxyphenylpyruvate dioxygenase. Arch Biochem Biophys 433:117–128
Nygard Y, Mojzita D, Toivari M, Penttila M, Wiebe MG, Ruohonen L (2014) The diverse role of Pdr12 in resistance to weak organic acids. Yeast 31:219–232
Oud B, van Maris AJA, Daran JM, Pronk JT (2012) Genome-wide analytical approaches for reverse metabolic engineering of industrially relevant phenotypes in yeast. FEMS Yeast Res 12:183–196
Oud B, Guadalupe-Medina V, Nijkamp JF, de Ridder D, Pronk JT, van Maris AJA, Daran JM (2013) Genome duplication and mutations in ACE2 cause multicellular, fast-sedimenting phenotypes in evolved Saccharomyces cerevisiae. Proc Natl Acad Sci USA 110:4223–4231
Patnaik R, Louie S, Gavrilovic V, Perry K, Stemmer WPC, Ryan CM, del Cardayre S (2002) Genome shuffling of Lactobacillus for improved acid tolerance. Nat Biotechnol 20:707–712
Pinel D, D’Aoust F, del Cardayre SB, Bajwa PK, Lee H, Martin VJJ (2011) Saccharomyces cerevisiae genome shuffling through recursive population mating leads to improved tolerance to spent sulfite liquor. Appl Environ Microbiol 77:4736–4743
Pinel D, Colatriano D, Jiang H, Lee H, Martin VJJ (2015) Deconstructing the genetic basis of spent sulfite liquor tolerance using deep sequencing of genome shuffled yeast. Biotech Biofuels 8:53. doi:10.1186/s13068-015-0241-z
Piper P, Mahe Y, Thompson S, Pandjaitan R, Holyoak C, Egner R, Muhlbauer M, Coote P, Kuchler K (1998) The Pdr12 ABC transporter is required for the development of weak organic acid resistance in yeast. EMBO J 17:4257–4265
Quan XX, Tsoulos P, Kuritzky A, Zhang R, Stochaj U (2006) The carrier Msn5p/Kap142p promotes nuclear export of the hsp70 Ssa4p and relocates in response to stress. Mol Microbiol 62:592–609
Rastogi RP, Kumar A, Tyagi MB, Sinha RP (2010) Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. J Nucleic Acids 2010:1–32
Reinhold R, Kruger V, Meinecke M, Schulz C, Schmidt B, Grunau SD, Guiard B, Wiedemann N, van der Laan M, Wagner R, Rehling P, Dudek J (2012) The channel-forming Sym1 protein is transported by the TIM23 complex in a presequence-independent manner. Mol Cell Biol 32:5009–5021
Richardson TL, Harner NK, Bajwa PK, Trevors JT, Lee H (2011) Approaches to deal with toxic inhibitors during fermentation of lignocellulosic substrates. In: Zhu JJY, Zhang X, Pan XJ (eds) Sustainable production of fuels, chemicals, and fibers from forest biomass. ACS Symposium Series; American Chemical Society, Washington, pp 171–202
Sá-Correia I, dos Santos SC, Teixeira MC, Cabrito TR, Mira NP (2009) Drug: H+ antiporters in chemical stress response in yeast. Trends Microbiol 17:22–31
Saavedra CA, Hammell CM, Heath CV, Cole CN (1997) Yeast heat shock mRNAs are exported through a distinct pathway defined by Rip1p. Genes Dev 11:2845–2856
Sasano Y, Watanabe D, Ukibe K, Inai T, Ohtsu I, Shimoi H, Takagi H (2012) Overexpression of the yeast transcription activator Msn2 confers furfural resistance and increases the initial fermentation rate in ethanol production. J Biosci Bioeng 113:451–455
Schauer A, Knauer H, Ruckenstuhl C, Fussi H, Durchschlag M, Potocnik U, Frohlich KU (2009) Vacuolar functions determine the mode of cell death. Biochem Biophys Acta 1793:540–545
Schneider H, Wang PY, Chan YK, Maleszka R (1981) Conversion of d-xylose into ethanol by the yeast Pachysolen tannophilus. Biotechnol Lett 3:82–92
Slininger PJ, Bothast RJ, Van Cauwenberge JE, Kurtzman CP (1982) Conversion of d-xylose to ethanol by the yeast Pachysolen tannophilus. Biotechnol Bioeng 24:371–384
Sugiyama M, Akase SP, Nakanishi R, Horie H, Kaneko Y, Harashima S (2014) Nuclear localization of Haa1, which is linked to its phosphorylation status, mediates lactic acid tolerance in Saccharomyces cerevisiae. Appl Environ Microbiol 80:3488–3495
Takahashi T, Shimoi H, Ito K (2001) Identification of genes required for growth under ethanol stress using transposon mutagenesis in Saccharomyces cerevisiae. Mol Genet Genomics 265:1112–1119
Tanaka K, Ishii Y, Ogawa J, Shima J (2012) Enhancement of acetic acid tolerance in Saccharomyces cerevisiae by overexpression of the HAA1 gene, encoding a transcriptional activator. Appl Environ Microbiol 78:8161–8163
Theander O (1991) Chemical analysis of lignocellulose materials. Anim Feed Sci Technol 32:35–44
Trott A, Morano KA (2004) SYM1 is the stress-induced Saccharomyces cerevisiae ortholog of the mammalian kidney disease gene Mpv17 and is required for ethanol metabolism and tolerance during heat shock. Eukaryot Cell 3:620–631
Warner JR (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24:437–440
Webb SR, Lee H (1990) Regulation of d-xylose utilization by hexoses in pentose-fermenting yeasts. Biotechnol Adv 8:685–697
Wright MB, Howell EA, Gaber RF (1996) Amino acid substitutions in membrane-spanning domains of Hol1, a member of the major facilitator superfamily of transporters, confer nonselective cation uptake in Saccharomyces cerevisiae. J Bacteriol 178:7197–7205
Acknowledgments
This research was supported by the NSERC Bioconversion Network and BioFuelNet. We thank Cletus Kurtzman (USDA ARS, Peoria, IL, USA) for providing P. t annophilus NRRL Y-2460, Frank Giust (Tembec Inc., Témiscaming, QC, Canada) for providing the HW SSL and Mike Rushton (formerly of Lignol, Burnaby, BC, Canada) for providing the aspen hydrolysate.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Harner, N.K., Bajwa, P.K., Formusa, P.A. et al. Determinants of tolerance to inhibitors in hardwood spent sulfite liquor in genome shuffled Pachysolen tannophilus strains. Antonie van Leeuwenhoek 108, 811–834 (2015). https://doi.org/10.1007/s10482-015-0537-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-015-0537-9