Abstract
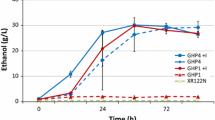
Lignocellulosic biomass conversion inhibitors, furfural and HMF, inhibit microbial growth and interfere with subsequent fermentation of ethanol, posing significant challenges for a sustainable cellulosic ethanol conversion industry. Numerous yeast genes were found to be associated with the inhibitor tolerance. However, limited knowledge is available about mechanisms of the tolerance and the detoxification of the biomass conversion inhibitors. Using a robust standard for absolute mRNA quantification assay and a recently developed tolerant ethanologenic yeast Saccharomyces cerevisiae NRRL Y-50049, we investigate pathway-based transcription profiles relevant to the yeast tolerance and the inhibitor detoxification. Under the synergistic inhibitory challenges by furfural and HMF, Y-50049 was able to withstand the inhibitor stress, in situ detoxify furfural and HMF, and produce ethanol, while its parental control Y-12632 failed to function till 65 h after incubation. The tolerant strain Y-50049 displayed enriched genetic background with significantly higher abundant of transcripts for at least 16 genes than a non-tolerant parental strain Y-12632. The enhanced expression of ZWF1 appeared to drive glucose metabolism in favor of pentose phosphate pathway over glycolysis at earlier steps of glucose metabolisms. Cofactor NAD(P)H generation steps were likely accelerated by enzymes encoded by ZWF1, GND1, GND2, TDH1, and ALD4. NAD(P)H-dependent aldehyde reductions including conversion of furfural and HMF, in return, provided sufficient NAD(P)+ for NAD(P)H regeneration in the yeast detoxification pathways. Enriched genetic background and a well maintained redox balance through reprogrammed expression responses of Y-50049 were accountable for the acquired tolerance and detoxification of furfural to furan methanol and HMF to furan dimethanol. We present significant gene interactions and regulatory networks involved in NAD(P)H regenerations and functional aldehyde reductions under the inhibitor stress.







Similar content being viewed by others
References
Almeida JRM, Roder A, Modig T, Laadan B, Liden G, Gorwa-Grauslund M (2008) NADH- versus NADPH-coupled reduction of 5-hydroxymethylfurfural HMF and its implications on product distribution in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 78:939–945
Antal MJ, Leesomboon T, Mok WS, Richards GN (1991) Mechanism of formation of 2-furaldehyde from d-xylose. Carbohydr Res 217:71–85
Applied Biosystem (2006) Amplification efficiency of TagMan gene expression assays. (Application Note Publication 127AP05-03) p 5
Baker SC, Bauer SR, Beyer RP, Brenton JD, Bromley B, Burrill J, Causton H, Conley MP, Elespuru R, Fero M, Foy C, Fuscoe J, Gao X, Gerhold DL, Gilles P, Goodsaid F, Guo X, Hackett J, Hockett RD, Ikonomi P, Irizarry RA, Kawasaki ES, Kaysser-Kranich T, Kerr K, Kiser G, Koch WH, Lee KY, Liu C, Liu ZL, Lucas A, Manohar CF, Miyada G, Modrusan Z, Parkes H, Puri RK, Reid L, Ryder TB, Salit M, Samaha RR, Scherf U, Sendera TJ, Setterquist RA, Shi L, Shippy R, Soriano JV, Wagar EA, Warrington JA, Williams M, Wilmer F, Wilson M, Wolber PK, Wu X, Zadro R (2005) The external RNA controls consortium: a progress report. Nat Methods 2:731–734
Banerjee N, Bhatnagar R, Viswanathan L (1981) Inhibition of glycolysis by furfural in Saccharomyces cerevisiae. Eur J Appl Microbiol Biotechnol 11:226–228
Bothast RJ, Saha BC (1997) Ethanol production from agricultural biomass substrate. Adv Appl Microbiol 44:261–286
Bro C, Regenberg B, Nielsen J (2004) Genome-wide transcriptional response of a Saccharomyces cerevisiae strain with an altered redox metabolism. Biotechnol Bioeng 85:269–276
Chung IS, Lee YY (1985) Ethanol fermentation of crude acid hydrolyzate of cellulose using high-level yeast inocula. Biotechnol Bioeng 27:308–315
Fisk DG, Ball CA, Dolinski K, Engel SR, Hong EL, Issel-Tarver L, Schwartz K, Sethuraman A, Botstein D, Cherry JM (2006) Saccharomyces cerevisiae S288C genome annotation: a working hypothesis. Yeast 23:857–865
Friedman N (2004) Inferring cellular networks using probabilistic graphical models. Science 303:799–805
Gorsich SW, Dien BS, Nichols NN, Slininger PJ, Liu ZL, Skory C (2006) Tolerance to furfural-induced stress is associated with pentose phosphate pathway genes ZWF1, GND1, RPE1, and TKL1 in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 71:339–349
Horvath IS, Franzen CJ, Taherzadeh MJ, Niklasson C, Liden G (2003) Effects of furfural on the respiratory metabolism of Saccharomyces cerevisiae in glucose-limited chemostats. Appl Environ Microbiol 69:4076–4086
Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M (2006) From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34:D354–D357
Khan QA, Hadi SA (1994) Inactivation and repair of bacteriophage lambda by furfural. Biochem Mol Biol Int 32:379–385
Klinke HB, Thomsen AB, Ahring BK (2004) Inhibition of ethanol-producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl Microbiol Biotechnol 66:10–26
Larsson S, Palmqvist E, Hahn-Hägerdal B, Tengborg C, Stenberg K, Zacchi G, Nilvebrant N (1999) The generation of fermentation inhibitors during dilute acid hydrolysis of softwood. Enzyme Microb Technol 24:151–159
Lewkowski J (2001) Synthesis, chemistry and applications of 5-hydroxymethylfurfural and its derivatives. Arkivoc 1:17–54
Liu ZL (2006) Genomic adaptation of ethanologenic yeast to biomass conversion inhibitors. Appl Microbiol Biotechnol 73:27–36
Liu ZL, Blaschek HP (2009) Lignocellulosic biomass conversion to ethanol by Saccharomyces. In: Vertes A, Qureshi N, Yukawa H, Blaschek H (eds) Biomass to biofuels. Wiley, West Sussex, pp 17–36
Liu W, Saint DA (2002) A new quantitative method of real time reverse transcription polymerase chain reaction assay based on simulation of polymerase chain reaction kinetics. Analytical Biochem 302:52–59
Liu ZL, Slininger PJ (2005) Development of genetically engineered stress tolerant ethanologenic yeasts using integrated functional genomics for effective biomass conversion to ethanol. In: Outlaw J, Collins K, Duffield J CAB International (eds) Agriculture as a producer and consumer of energy, Wallingford, UK, pp 283–294
Liu ZL, Slininger PJ (2006) Transcriptome dynamics of ethanologenic yeast in response to 5-hydroxymethylfurfural stress related to biomass conversion to ethanol. In: Mendez-Vilas A (ed) Modern multidisciplinary applied microbiology: exploiting microbes and their interactions. Wiley-VCH, Weinheim, pp 679–685
Liu ZL, Slininger PJ (2007) Universal external RNA controls for microbial gene expression analysis using microarray and qRT-PCR. J Microbiol Methods 68:486–496
Liu ZL, Slininger PJ, Dien BS, Berhow MA, Kurtzman CP, Gorsich SW (2004) Adaptive response of yeasts to furfural and 5-hydroxymethylfurfural and new chemical evidence for HMF conversion to 2,5-bis-hydroxymethylfuran. J Ind Microbiol Biotechnol 31:345–352
Liu ZL, Slininger PJ, Gorsich SW (2005) Enhanced biotransformation of furfural and 5-hydroxymethylfurfural by newly developed ethanologenic yeast strains. Appl Biochem Biotechnol 121–124:451–460
Liu ZL, Saha BC, Slininger PJ (2008a) Lignocellulosic biomass conversion to ethanol by Saccharomyces. In: Wall J, Harwood C, Demain A (eds) Bioenergy. ASM Press, Washington DC, pp 17–36
Liu ZL, Moon J, Andersh AJ, Slininger PJ, Weber S (2008b) Multiple gene mediated NAD(P)H-dependent aldehyde reduction is a mechanism of in situ detoxification of furfural and HMF by ethanologenic yeast Saccharomyces cerevisiae. Appl Microbiol Biotechnol 81:743–753
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Luo C, Brink D, Blanch HW (2002) Identification of potential fermentation inhibitors in conversion of hybrid poplar hydrolyzate to ethanol. Biomass Bioenergy 22:125–138
Modig T, Liden G, Taherzadeh MJ (2002) Inhibition effects of furfural on alcohol dehydrogenase, aldehyde dehydrogenase and pyruvate dehydrogenase. Biochem J 363:769–776
Morimoto S, Murakami M (1967) Studies on fermentation products from aldehyde by microorganisms: the fermentative production of furfural alcohol from furfural by yeasts (part I). J Ferm Technol 45:442–446
Nemirovskii V, Gusarova L, Rakhmilevich Y, Sizov A, Kostenko V (1989) Pathways of furfurol and oxymethyl furfurol conversion in the process of fodder yeast cultivation. Biotekhnologiia 5:285–289
Nilsson A, Gorwa-Grauslund MF, Hahn-Hagerdal B, Liden G (2005) Cofactor dependence in furan reduction by Saccharomyces cerevisiae in fermentation of acid-hydrolyzed lignocellulose. Appl Environ Microbiol 71:7866–7871
Palmqvist E, Almeida JS, Hahn-Hagerdal B (1999) Influence of furfural on anaerobic glycolytic kinetics of Saccharomyces cerevisiae in batch culture. Biotechnol Bioeng 62:447–454
Petersson A, Almeida JR, Modig T, Karhumma K, Hahn-Hägerdal B, Gorwa-Grauslund MF (2006) A 5-hydroxymethylfurfural reducing enzyme encoded by the Saccharomyces cerevisiae ADH6 gene conveys HMF tolerance. Yeast 23:455–464
R Development Core Team (2008) R: a language and environment for statistical computing. R foundation for statistical computing, Vienna, Austria, http://www.R-project.org. ISBN 3-900051-07-0
Rozen S, Skaletsky H (2000) Bioinformatics methods and protocols. In: Krawetz S, Misener S (eds) Methods in molecular biology. Humana Press, Totowa, pp 365–386
Sanchez B, Bautista J (1988) Effects of furfural and 5-Hydroxymethylfurfrual on the fermentation of Saccharomyces cerevisiae and biomass production from Candida guilliermondii. Enzyme Microb Technol 10:315–318
Song M, Liu ZL (2007) A linear discrete dynamic system model for temporal gene interaction and regulatory network influence in response to bioethanol conversion inhibitor HMF for ethanologenic yeast. Lect Notes Bioinfomatics 4532:77–95
Song M, Ouyang Z, Liu ZL (2009) Discrete dynamic system modeling for gene regulatory networks of HMF tolerance for ethanologenic yeast. IET Sys Biology 3:203–218
Taherzadeh MJ, Gustafsson L, Niklasson C, Liden G (1999) Conversion of furfural in aerobic and anaerobic batch fermentation of glucose by Saccharomyces cerevisiae. J Biosci Bioeng 87:169–174
Taherzadeh MJ, Gustafsson L, Niklasson C, Liden G (2000) Physiological effects of 5-hydroxymethylfurfural on Saccharomyces cerevisiae. Appl Microbiol Biotechnol 53:701–708
The External RNA Control Consortium (2005) Proposed methods for testing and selecting ERCC external RNA controls. BMC Genomics 6:150
Villa GP, Bartroli R, Lopez R, Guerra M, Enrique M, Penas M, Rodriquez E, Redondo D, Iglesias I, Diaz I (1992) Microbial transformation of furfural to furfuryl alcohol by Saccharomyces cerevisiae. Acta Biotechnol 12:509–512
Acknowledgments
We thank Scott Weber and Stephanie Thompson for technical assistance and Pat Slininger for discussions. This work was supported by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant number 2006-35504-17359.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
The mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lewis Liu, Z., Ma, M. & Song, M. Evolutionarily engineered ethanologenic yeast detoxifies lignocellulosic biomass conversion inhibitors by reprogrammed pathways. Mol Genet Genomics 282, 233–244 (2009). https://doi.org/10.1007/s00438-009-0461-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-009-0461-7