Abstract
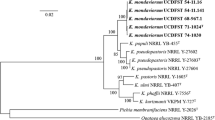
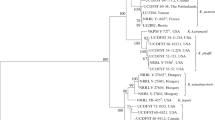
A novel methanol assimilating yeast species Komagataella kurtzmanii is described using the type strain VKPM Y-727 (=KBP Y-2878 = UCD-FST 76-20 = Starmer #75-208.2 = CBS 12817 = NRRL Y-63667) isolated by W.T. Starmer from a fir flux in the Catalina Mountains, Southern AZ, USA. The new species is registered in MycoBank under MB 803919. The species was differentiated by divergence in gene sequences for D1/D2 LSU rRNA, ITS1-5.8S-ITS2, RNA polymerase subunit I, translation elongation factor-1α and mitochondrial small subunit rRNA. K. kurtzmanii differs from its phenotypically similar sibling species Komagataella pastoris, Komagataella pseudopastoris, Komagataella phaffii, Komagataella populi and Komagataella ulmi by absence of growth at 35 °C and inability to assimilate trehalose.


Similar content being viewed by others
References
Chen MT, Lin S, Shandil I, Andrews D, Stadheim TA, Choi BK (2012) Generation of diploid Pichia pastoris strains by mating and their application for recombinant protein production. Microb Cell Factories 11:91
Cregg JM, Madden KR (1988) Development of the methylotrophic yeast, Pichia pastoris, as a host system for the production of foreign proteins. Dev Ind Microbiol 29:33–41
Cregg JM, Vedvick TS, Raschke WC (1993) Recent advances in the expression of foreign genes in Pichia pastoris. Biotechnology 11:905–910
Cregg JM, Latham J, Litton M, Schatzman R, Tolstorukov II (2012) Methods of synthesizing heteromultimeric polypeptides in yeast using a haploid mating strategy. US Patent 8.268.582
Dlauchy D, Tornai-Lehoczki J, Fülöp L, Péter G (2003) Pichia (Komagataella) pseudopastoris sp. nov., a new yeast species from Hungary. Antonie Van Leeuwenhoek 83:327–332
Kudrjawzev WI (1960) Die systematic der Hefen. Academic Verlag, Berlin
Kurtzman CP (1984) Synonymy of the yeast genera Hansenula and Pichia demonstrated through comparisons of deoxyribonucleic acid relatedness. Antonie Van Leeuwenhoek 50:209–217
Kurtzman CP (1998) Pichia E.C. Hansen emend. Kurtzman. In: Kurtzman CP, Fell JW (eds) The yeasts, a taxonomic study, 4th edn. Elsevier, Amsterdam, pp 273–352
Kurtzman CP (2005) Description of Komagataella phaffii sp. nov. and the transfer of Pichia pseudopastoris to the methylotrophic yeast genus Komagataella. Int J Syst Evol Microbiol 55:973–976
Kurtzman CP (2009) Biotechnological strains of Komagataella (Pichia) pastoris are Komagataella phaffii as determined from multigene sequence analysis. J Ind Microbiol Biotechnol 36:1435–1438
Kurtzman CP (2011) Komagataella Y. Yamada, Matsuda, Maeda, Mikata (1995). In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier Science B.V, Amsterdam, pp 491–495
Kurtzman CP (2012) Komagataella populi sp. nov. and Komagataella ulmi sp. nov., two new methanol assimilating yeasts from exudates of deciduous trees. Antonie Van Leeuwenhoek 101:859–868
Kurtzman CP, Robnett CJ (1998) Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73:331–371
Kurtzman CP, Robnett CJ (2003) Phylogenetic relationships among yeasts of the “Saccharomyces complex” determined from multigene sequence analyses. FEMS Yeast Res 3:417–432
Kurtzman CP, Robnett CJ, Basehoar-Powers E (2008) Relationships among species of Pichia, Issatchenkia and Williopsis determined from multigene phylogenetic analysis and the proposal of Barnettozyma gen. nov., Lindnera gen. nov. and Wickerhamomyces gen. nov. FEMS Yeast Res 8:939–954
Kurtzman CP, Fell JW, Boekhout T, Robert V (2011) Methods for isolation, phenotypic characterization and maintenance of yeasts. In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier Science B.V, Amsterdam, pp 87–110
Macauley-Patrick S, Fazenda ML, McNeil B, Harvey LM (2005) Heterologous protein production using the Pichia pastoris expression system. Yeast 22:249–270
McClary DD, Nulty WL, Miller GR (1959) Effect of potassium versus sodium in the sporulation of Saccharomyces. J Bacteriol 78:362–368
Naumov GI (1979) Genetic concept of genus in fungi. Dokl Biol Sci 241(4):345–347
Naumov GI (1986) Genosystematics of yeast genus Zygofabospora Kudriavzev emend. G. Naumov. Mol Genet Microbiol Virol (Moscow) 5:10–14
Naumov GI (1987a) Genetic basis for classification and identification of the ascomycetous yeasts. Stud Mycol 30:469–475
Naumov GI (1987b) Results of the genosystematics of the yeast Williopsis Zender and Zygowilliopsis Kudriavzev. Mol Genet Microbiol Virol (Moscow) 2:1–7
Naumov GI (1988) Genus as a genetic system. In: Aspects of microevolution. Nauka, Moscow, pp 112–113 (in Russian)
Naumov GI (1996) Genetic identification of biological species in the Saccharomyces sensu stricto complex. J Ind Microbiol 17:295–302
Naumov GI, Naumova ES (2002) Five new combinations in the yeast genus Zygofabospora Kudriavzev emend. G. Naumov (pro parte Kluyveromyces) based on genetic data. FEMS Yeast Res 2:39–46
Naumov GI, Naumova ES, Kondratieva VI, Bulat SA, Mironenko NV, Mendonça-Hagler LC, Hagler AN (1997) Genetic and molecular delineation of three sibling species in the Hansenula polymorpha complex. Syst Appl Microbiol 20:50–56
Naumov GI, James SA, Naumova ES, Louis EJ, Roberts IN (2000) Three new species in the Saccharomyces sensu stricto complex: Saccharomyces cariocanus, Saccharomyces kudriavzevii and Saccharomyces mikatae. Int J Syst Evol Microbiol 50:193–1942
Naumov GI, Naumova ES, Smith MT, de Hoog GS (2006) Molecular-genetic diversity of the genus Arthroascus: Arthroascus babieviae sp.nov., Arthroascus fermentans var. arxii nov. var. and geographic populations of Arthroascus schoenii. Int J Syst Evol Microbiol 56:1997–2007
Naumov GI, Kondratieva VI, Naumova ES (2009) Taxonomic genetics of Zygowilliopsis yeasts. Russ J Genet 45:1422–1427
Naumov GI, Naumova ES, Masneuf-Pomarède I (2010) Genetic identification of new biological species Saccharomyces arboricolus Wang et Bai. Antonie Van Leeuwenhoek 98:1–7
Naumova ES, Smith MTh, Boekhout T, Hoog GS, Naumov GI (2001) Molecular differentiation of sibling species in the Galactomyces geotrichum complex. Antonie Van Leeuwenhoek 80:263–273
Scheda R, Yarrow D (1966) The instability of physiological properties used as criteria in the taxonomy of yeasts. Arch Mikrobiol 55:209–225
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding Consortium (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci (USA) 109:6241–6246
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Wegner EH (1983) Biochemical conversions by yeast fermentation at high-cell densities. US Patent 44114329
Wu Q, James SA, Roberts IN, Moulton V, Huber KT (2008) Exploring contradictory phylogenetic relations in yeasts. FEMS Yeast Res 8:641–650
Yamada Y, Matsuda M, Maeda K, Mikata K (1995) The phylogenetic relationships of methanol-assimilating yeasts based on the partial sequences of 18S and 26S ribosomal RNAs: the proposal of Komagataella gen. nov. (Saccharomycetaceae). Biosci Biotechnol Biochem 59:439–444
Yarrow D (1998) Methods for the isolation, maintenance and identification of yeasts. In: Kurtzman CP, Fell JW (eds) The yeasts, a taxonomic study, 4th edn. Elsevier, Amsterdam, pp 77–100
Acknowledgments
Synthesis of oligonucleotide primers and sequencing of the D1/D2 26S rRNA, translation elongation factor-1α, RNA polymerase II, mitochondrial small subunit rRNA and ITS1-5.8S-ITS2 were performed using the equipment of the Centre for Collective Use of GosNIIgenetika (Moscow).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Naumov, G.I., Naumova, E.S., Tyurin, O.V. et al. Komagataella kurtzmanii sp. nov., a new sibling species of Komagataella (Pichia) pastoris based on multigene sequence analysis. Antonie van Leeuwenhoek 104, 339–347 (2013). https://doi.org/10.1007/s10482-013-9956-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-013-9956-7