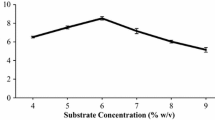
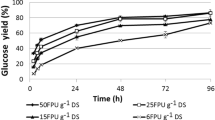
Abstract
The aim of this study was a comprehensive analysis of the effects of the component enzymes of cellulase derived from Trichoderma reesei strain PC-3-7 on biomass saccharification. We used cellulases with deleted CBH I, CBH II, or EG I, which contain all other component enzymes, for saccharification of differently pretreated biomasses of rice straw, Erianthus, eucalyptus, and Japanese cedar. We found that CBH I was the most effective in saccharification of all pretreated cellulosic biomasses, although the effect was weaker in saccharification of sulfuric acid- and hydrothermally pretreated rice straw than of others; CBH II was more effective for rice straw than for eucalyptus, and was the most effective at the early stages of biomass degradation; EG I had little effect on pretreated biomasses, in particular, it had no effect on steam-exploded Japanese cedar. Thus, the effects of the main component enzymes depend on the biomass source and pretreatment. These findings will likely help to improve cellulase for industrial use.





Similar content being viewed by others
References
Balat M, Balat H (2009) Recent trends in global production and utilization of bio-ethanol fuel. Appl Energ 86:2273–2282
Banerjee G, Car S, Scott-Craig JS, Borrusch MS, Bongers M, Walton JD (2010) Synthetic multi-component enzyme mixtures for deconstruction of lignocellulosic biomass. Bioresour Biotechnol 101:9097–9105
Barr BK, Hsieh YL, Ganem B, Wilson DB (1996) Identification of two functionally different classes of exocellulases. Biochemistry-US 35:586–592
Billard H, Faraj A, Lopes Ferreira N, Menir S, Heiss-Blanquet S (2012) Optimization of a synthetic mixture composed of major Trichoderma reesei enzymes for the hydrolysis of steam-exploded wheat straw. Biotechnol Biofuels 5:9
Boer H, Teeri TT, Koivula A (2000) Characterization of Trichoderma reesei cellobiohydrolase Cel7A secreted from Pichia pastoris using two different promoters. Biotechnol Bioeng 69:489–494
Boisset C, Fraschini C, Schülein M, Henrissat B, Chanzy H (2000) Imaging the enzymatic digestion of bacterial cellulose ribbons reveals the endo character of the cellobiohydrolase Cel6A from Humicola insolens and its mode of synergy with cellobiohydrolase Cel7A. Appl Environ Microb 66:1444–1452
Chen H, Hayn M, Esterbauer H (1992) Purification and characterization of two extracellular beta-glucosidases from Trichoderma reesei. Biochim Biophys Acta 1121:54–60
Chirico WJ, Brown RD Jr (1987) β-glucosidase from Trichoderma reesei. Substate-binding region and mode of action on [1-3H] cello-oligosaccharides. Eur J Biochem 165:343–351
Donaldson LA, Wong KKY, Mackie KL (1988) Ultrastructure of steam-exploded wood. Wood Sci Technol 22:103–114
Elazzouzi-Hafraoui S, Nishiyama Y, Putaux JL, Heux L, Dubreuil F, Rochas C (2008) The shape and size distribution of crystalline nanoparticles prepared by acid hydrolysis of native cellulose. Biomacromolecules 9:57–65
Eriksson T, Karlsson J, Tjerneld F (2002) A model explaining declining rate in hydrolysis of lignocellulose substrates with cellobiohydrolase I (Cel7A) and endoglucanase I (Cel7B) of Trichoderma reesei. Appl Biochem Biotechnol 101:41–60
Herpoël-Gimbert I, Margeot A, Dolla A, Jan G, Mollé D, Lignon S, Mathis H, Sigoillot J, Monot F, Asther M (2008) Comparative secretome analyses of two Trichoderma reesei RUT-C30 and CL847 hypersecretory strains. Biotechnol Biofuels 1:18
Kabel MA, Maarel MJEC, Klip G, Voragen AGJ, Schols HA (2006) Standard assays do not predict the efficiency of commercial cellulase preparations towards plant materials. Biotechnol Bioeng 93:56–63
Kawai T, Nakazawa H, Ida N, Okada H, Tani S, Sumitani J, Kawaguchi T, Ogasawara W, Morikawa Y, Kobayashi Y (2012) Analysis of the saccharification capability of high-functional cellulase JN11 for various pretreated biomasses through a comparison with commercially available counterparts. J Ind Microbiol Biotechnol 39:1741–1749
Kawamori M, Morikawa Y, Takasawa S (1986) Induction and production of cellulases by L-sorbose in Trichoderma reesei. Appl Microbiol Biotechnol 24:449–453
Kipper K, Väljamäe P, Johansson G (2005) Processive action of cellobiohydrolase Cel7A from Trichoderma reesei is revealed as ‘burst’ kinetics on fluorescent polymeric model substrates. Biochem J 385:527–535
Kleman-Leyer KM, Siika-Aho M, Teeri TT, Kirk TK (1996) The Cellulases endoglucanase I and cellobiohydrolase II of Trichoderma reesei act synergistically to solubilize native cotton cellulose but not to decrease its molecular size. Appl Environ Microb 62:2883–2887
Mach RL, Zeilinger S (2003) Regulation of gene expression in industrial fungi: Trichoderma. Appl Microbiol Biotechnol 60:515–522
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428
Nagendran S, Hallen-Adams HE, Paper JM, Aslam N, Walton JD (2009) Reduced genomic potential for secreted plant cell-wall-degrading enzymes in the ectomycorrhizal fungus Amanita bisporigera, based on the secretome of Trichoderma reesei. Fungal Genet Biol 46:427–435
Nakazawa H, Kawai T, Ida N, Shida Y, Kobayashi Y, Okada H, Tani S, Sumitani J, Kawaguchi T, Morikawa Y, Ogasawara W (2011) Construction of a recombinant Trichoderma reesei strain expressing Aspergillus aculeatus β-glucosidase 1 for efficient biomass conversion. Biotechnol Bioeng 109:92–99
Nogawa M, Goto M, Okada H, Morikawa Y (2001) l-Sorbose induces cellulase gene transcription in the cellulolytic fungus Trichoderma reesei. Curr Genet 38:329–334
Quinlan RJ, Sweeney MD, Leggio LL, Otten H, Poulsen JCN, Johansen KS, Krogh KBRM, Jørgensen CI, Tovborg M, Anthonsen A, Tryfona T, Walter CP, Dupree P, Xu F, Davies GJ, Walton PH (2011) Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. PNAS 108:15079–15084
Rahman Z, Shida Y, Furukawa T, Suzuki Y, Okada H, Ogasawara W, Morikawa Y (2009) Evaluation and characterization of Trichoderma reesei cellulase and xylanase promoters. Appl Microbiol Biotechnol 82:899–908
Rosgaard L, Pedersen S, Langston J, Akerhielm D, Cherry JR, Meyer AS (2007) Evaluation of minimal Trichoderma reesei cellulase mixtures on differently pretreated barley straw substrates. Biotechnol Progr 23:1270–1276
Saloheimo M, Paloheimo M, Hakola S, Pere J, Swanson B, Nyyssönen E, Bhatia A, Ward M, Penttilä M (2002) Swollenin, a Trichoderma reesei protein with sequence similarity to the plant expansins, exhibits disruption activity on cellulosic materials. Eur J Biochem 269:4202–4211
Silverstein RA, Chen Y, Sharma-Shivappa RR, Boyette MD, Osborne J (2007) A comparison of chemical pretreatment methods for improving saccharification of cotton stalks. Bioresour Technol 98:3000–3011
Sluiter A, Hames B, Ruiz R, Scarlata C, Sluiter J, Templeton D, Crocker D (2008) Determination of structural carbohydrates and lignin in biomass. NREL laboratory analytical procedures. http://www.eere.energy.gov/biomass/analytical_procedures.html#LAP-002
Suominen PL, Mäntylä AL, Karhunen T, Hakola S, Nevalainen H (1993) High frequency one-step gene replacement in Trichoderma reesei. II. Effects of deletions of individual cellulase genes. Mol Gen Genet 241:523–530
Vlasenko E, Schülein M, Cherry J, Xu F (2010) Substrate specificity of family 5, 6, 7, 9, 12, and 45 endoglucanases. Bioresour Technol 101:2405–2411
Zhang Y (2008) Reviving the carbohydrate economy via multi-product lignocellulose biorefineries. J Ind Microbiol Biotechnol 35:367–375
Zhao Y, Wu B, Yan B, Gao P (2004) Mechanism of cellobiose inhibition in cellulose hydrolysis by cellobiohydrolase. Sci China Ser C 47:18–24
Acknowledgments
This work was supported by a grant from the New Energy and Industrial Technology Development Organization (NEDO) Project.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kawai, T., Nakazawa, H., Ida, N. et al. A comprehensive analysis of the effects of the main component enzymes of cellulase derived from Trichoderma reesei on biomass saccharification. J Ind Microbiol Biotechnol 40, 805–810 (2013). https://doi.org/10.1007/s10295-013-1290-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-013-1290-6